 |
PDBsum entry 5tk5

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5tk5
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.6
- 3-hydroxyanthranilate 3,4-dioxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
(later stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-hydroxyanthranilate + O2 = (2Z,4Z)-2-amino-3-carboxymuconate 6-semialdehyde
|
 |
 |
 |
 |
 |
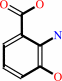 3-hydroxyanthranilate
3-hydroxyanthranilate
|
+
|
 O2
O2
|
=
|
(2Z,4Z)-2-amino-3-carboxymuconate 6-semialdehyde
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr D Struct Biol
73:340-348
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of human 3-hydroxyanthranilate 3,4-dioxygenase with native and non-native metals bound in the active site.
|
|
L.S.Pidugu,
H.Neu,
T.L.Wong,
E.Pozharski,
J.L.Molloy,
S.L.Michel,
E.A.Toth.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
3-Hydroxyanthranilate 3,4-dioxygenase (3HAO) is an enzyme in the microglial
branch of the kynurenine pathway of tryptophan degradation. 3HAO is a non-heme
iron-containing, ring-cleaving extradiol dioxygenase that catalyzes the addition
of both atoms of O2 to the kynurenine pathway metabolite
3-hydroxyanthranilic acid (3-HANA) to form quinolinic acid (QUIN). QUIN is a
highly potent excitotoxin that has been implicated in a number of
neurodegenerative conditions, making 3HAO a target for pharmacological
downregulation. Here, the first crystal structure of human 3HAO with the native
iron bound in its active site is presented, together with an additional
structure with zinc (a known inhibitor of human 3HAO) bound in the active site.
The metal-binding environment is examined both structurally and via inductively
coupled plasma mass spectrometry (ICP-MS), X-ray fluorescence spectroscopy (XRF)
and electron paramagnetic resonance spectroscopy (EPR). The studies identified
Met35 as the source of potential new interactions with substrates and
inhibitors, which may prove useful in future therapeutic efforts.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

