 |
PDBsum entry 5tic
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
X-ray structure of wild-type e. Coli acyl-coa thioesterase i at ph 5
|
|
Structure:
|
 |
Acyl-coa thioesterase i. Chain: a, b. Fragment: unp residues 28-208. Synonym: lecithinase b,lysophospholipase l1,protease i. Engineered: yes
|
|
Source:
|
 |
Escherichia coli. Organism_taxid: 562. Gene: tesa, apea, pldc, b0494, jw0483. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.65Å
|
R-factor:
|
0.188
|
R-free:
|
0.222
|
|
|
Authors:
|
 |
J.B.Thoden,H.M.Holden,M.J.Grisewood,N.J.Hernandez Lozada,N.P.Gifford, D.Mendez-Perez,H.A.Schoenberger,M.F.Allan,B.F.Pfleger,C.D.Marines
|
|
Key ref:
|
 |
M.J.Grisewood
et al.
(2017).
Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal,
7,
3837-3849.
PubMed id:
|
 |
|
Date:
|
 |
|
02-Oct-16
|
Release date:
|
26-Apr-17
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0ADA1
(TESA_ECOLI) -
Thioesterase 1/protease 1/lysophospholipase L1 from Escherichia coli (strain K12)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
208 a.a.
177 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.1.2
- arylesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a phenyl acetate + H2O = a phenol + acetate + H+
|
 |
 |
 |
 |
 |
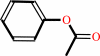 phenyl acetate
phenyl acetate
|
+
|
H2O
|
=
|
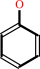 phenol
phenol
|
+
|
 acetate
acetate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.1.5
- lysophospholipase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1-acyl-sn-glycero-3-phosphocholine + H2O = sn-glycerol 3-phosphocholine + a fatty acid + H+
|
 |
 |
 |
 |
 |
 1-acyl-sn-glycero-3-phosphocholine
1-acyl-sn-glycero-3-phosphocholine
|
+
|
H2O
|
=
|
sn-glycerol 3-phosphocholine
|
+
|
 fatty acid
fatty acid
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.2.14
- oleoyl-[acyl-carrier-protein] hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(9Z)-octadecenoyl-[ACP] + H2O = (9Z)-octadecenoate + holo-[ACP] + H+
|
 |
 |
 |
 |
 |
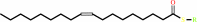 Oleoyl-[acyl-carrier-protein]
Oleoyl-[acyl-carrier-protein]
|
+
|
H(2)O
|
=
|
 [acyl-carrier-protein]
[acyl-carrier-protein]
|
+
|
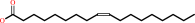 oleate
oleate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.3.1.2.2
- palmitoyl-CoA hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hexadecanoyl-CoA + H2O = hexadecanoate + CoA + H+
|
 |
 |
 |
 |
 |
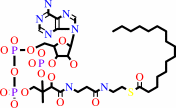 hexadecanoyl-CoA
hexadecanoyl-CoA
|
+
|
H2O
|
=
|
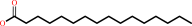 hexadecanoate
hexadecanoate
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
ACS Catal
7:3837-3849
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
|
|
M.J.Grisewood,
N.J.Hernandez Lozada,
J.B.Thoden,
N.P.Gifford,
D.Mendez-Perez,
H.A.Schoenberger,
M.F.Allan,
M.E.Floy,
R.Y.Lai,
H.M.Holden,
B.F.Pfleger,
C.D.Maranas.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Enzyme and metabolic engineering offer the potential to develop biocatalysts for
converting natural resources into a wide range of chemicals. To broaden the
scope of potential products beyond natural metabolites, methods of engineering
enzymes to accept alternative substrates and/or perform novel chemistries must
be developed. DNA synthesis can create large libraries of enzyme-coding
sequences, but most biochemistries lack a simple assay to screen for promising
enzyme variants. Our solution to this challenge is structure-guided mutagenesis
in which optimization algorithms select the best sequences from libraries based
on specified criteria (i.e. binding selectivity). Here, we demonstrate this
approach by identifying medium-chain (C6-C12) acyl-ACP
thioesterases through structure-guided mutagenesis. Medium-chain fatty acids,
products of thioesterase-catalyzed hydrolysis, are limited in natural abundance
compared to long-chain fatty acids; the limited supply leads to high costs of
C6-C10oleochemicals such as fatty alcohols, amines, and
esters. Here, we applied computational tools to tune substrate binding to the
highly-active 'TesA thioesterase inEscherichia coli.We used the IPRO
algorithm to design thioesterase variants with enhanced C12- or
C8-specificity while maintaining high activity. After four rounds of
structure-guided mutagenesis, we identified three thioesterases with enhanced
production of dodecanoic acid (C12) and twenty-seven thioesterases
with enhanced production of octanoic acid (C8). The top variants
reached up to 49% C12and 50% C8while exceeding native
levels of total free fatty acids. A comparably sized library created by random
mutagenesis failed to identify promising mutants. The chain length-preference of
'TesA and the best mutant were confirmedin vitrousing acyl-CoA
substrates. Molecular dynamics simulations, confirmed by resolved crystal
structures, of 'TesA variants suggest that hydrophobic forces govern 'TesA
substrate specificity. We expect that the design rules we uncovered and the
thioesterase variants identified will be useful to metabolic engineering
projects aimed at sustainable production of medium-chain oleochemicals.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

