 |
PDBsum entry 5tcf
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of tryptophan synthase from m. Tuberculosis - ligand-free form
|
|
Structure:
|
 |
Tryptophan synthase alpha chain. Chain: a, g, e, c. Engineered: yes. Tryptophan synthase beta chain. Chain: b, h, f, d. Fragment: unp 13-422. Engineered: yes. Other_details: lys101 is attached to plp
|
|
Source:
|
 |
Mycobacterium tuberculosis (strain atcc 25618 / h37rv). Organism_taxid: 83332. Strain: atcc 25618 / h37rv. Gene: trpa, rv1613, mtcy01b2.05. Expressed in: escherichia coli. Expression_system_taxid: 469008. Gene: trpb, rv1612, mtcy01b2.04.
|
|
Resolution:
|
 |
|
2.46Å
|
R-factor:
|
0.176
|
R-free:
|
0.207
|
|
|
Authors:
|
 |
K.Michalska,N.Maltseva,R.Jedrzejczak,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
S.Wellington
et al.
(2017).
A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat Chem Biol,
13,
943-950.
PubMed id:
|
 |
|
Date:
|
 |
|
15-Sep-16
|
Release date:
|
31-May-17
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, G, H, E, F, C, D:
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
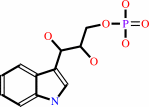 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
L-serine
Bound ligand (Het Group name = )
matches with 75.00% similarity
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
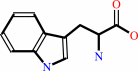 L-tryptophan
L-tryptophan
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Nat Chem Biol
13:943-950
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
|
|
S.Wellington,
P.P.Nag,
K.Michalska,
S.E.Johnston,
R.P.Jedrzejczak,
V.K.Kaushik,
A.E.Clatworthy,
N.Siddiqi,
P.McCarren,
B.Bajrami,
N.I.Maltseva,
S.Combs,
S.L.Fisher,
A.Joachimiak,
S.L.Schreiber,
D.T.Hung.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
New antibiotics with novel targets are greatly needed. Bacteria have numerous
essential functions, but only a small fraction of such processes-primarily those
involved in macromolecular synthesis-are inhibited by current drugs. Targeting
metabolic enzymes has been the focus of recent interest, but effective
inhibitors have been difficult to identify. We describe a synthetic azetidine
derivative, BRD4592, that kills Mycobacterium tuberculosis (Mtb) through
allosteric inhibition of tryptophan synthase (TrpAB), a previously untargeted,
highly allosterically regulated enzyme. BRD4592 binds at the TrpAB α-β-subunit
interface and affects multiple steps in the enzyme's overall reaction, resulting
in inhibition not easily overcome by changes in metabolic environment. We show
that TrpAB is required for the survival of Mtb and Mycobacterium marinum in vivo
and that this requirement may be independent of an adaptive immune response.
This work highlights the effectiveness of allosteric inhibition for targeting
proteins that are naturally highly dynamic and that are essential in vivo,
despite their apparent dispensability under in vitro conditions, and suggests a
framework for the discovery of a next generation of allosteric inhibitors.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

