 |
PDBsum entry 5sfh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Hydrolase/hydrolase inhibitor
|
PDB id
|
|

|

|
5sfh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase/hydrolase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of human phosphodiesterase 10 in complex with n(c1cc1)(c)c2nn(c(n2)ccc4nn3c(cnc(c3n4)c)c)c, micromolar ic50=0.042255
|
|
Structure:
|
 |
Camp and camp-inhibited cgmp 3',5'-cyclic phosphodiesterase 10a. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pde10a. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.29Å
|
R-factor:
|
0.176
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
C.Joseph,A.Flohr,J.Benz,D.Schlatter,M.G.Rudolph
|
|
Key ref:
|
 |
A.Tosstorff
et al.
(2022).
A high quality, Industrial data set for binding affin prediction: performance comparison in different early discovery scenarios..
J comput aided mol des,
36,
753.
PubMed id:
|
 |
|
Date:
|
 |
|
21-Jan-22
|
Release date:
|
12-Oct-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9Y233
(PDE10_HUMAN) -
cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1055 a.a.
313 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.17
- 3',5'-cyclic-nucleotide phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a nucleoside 3',5'-cyclic phosphate + H2O = a nucleoside 5'-phosphate + H+
|
 |
 |
 |
 |
 |
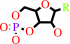 nucleoside 3',5'-cyclic phosphate
nucleoside 3',5'-cyclic phosphate
|
+
|
H2O
|
=
|
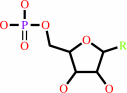 nucleoside 5'-phosphate
nucleoside 5'-phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

