 |
PDBsum entry 5ryh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Inpp5d pandda analysis group deposition -- crystal structure of the phosphatase and c2 domains of ship1 in complex with z1212984951
|
|
Structure:
|
 |
Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1. Chain: a. Synonym: inositol polyphosphate-5-phosphatase d,inositol polyphosphate-5-phosphatase of 145 kda,sip-145,phosphatidylinositol 4,5-bisphosphate 5-phosphatase,sh2 domain-containing inositol 5'- phosphatase 1,ship-1,p150ship,hp51cn. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: inpp5d, ship, ship1. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108
|
|
Resolution:
|
 |
|
1.72Å
|
R-factor:
|
0.196
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
W.J.Bradshaw,J.A.Newman,F.Von Delft,C.H.Arrowsmith,A.M.Edwards, C.Bountra,O.Gileadi
|
|
Key ref:
|
 |
W.J.Bradshaw
et al.
Inpp5d pandda analysis group deposition.
To be published,
.
PubMed id:
|
 |
|
Date:
|
 |
|
30-Oct-20
|
Release date:
|
11-Nov-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q92835
(SHIP1_HUMAN) -
Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1189 a.a.
460 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.36
- phosphoinositide 5-phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
1-Phosphatidyl-myo-inositol Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate) + H2O = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol 4-phosphate) + phosphate
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-4,5-bisphosphate)
|
+
|
H2O
|
=
|
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol 4-phosphate)
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.56
- inositol-polyphosphate 5-phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
1D-myo-inositol 1,4,5-trisphosphate + H2O = 1D-myo-inositol 1,4- bisphosphate + phosphate
|
|
2.
|
1D-myo-inositol 1,3,4,5-tetrakisphosphate + H2O = 1D-myo-inositol 1,3,4-trisphosphate + phosphate
|
|
 |
 |
 |
 |
 |
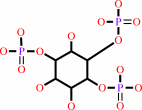 1D-myo-inositol 1,4,5-trisphosphate
1D-myo-inositol 1,4,5-trisphosphate
|
+
|
H2O
|
=
|
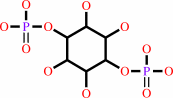 1D-myo-inositol 1,4- bisphosphate
1D-myo-inositol 1,4- bisphosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 1D-myo-inositol 1,3,4,5-tetrakisphosphate
1D-myo-inositol 1,3,4,5-tetrakisphosphate
|
+
|
H2O
|
=
|
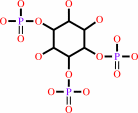 1D-myo-inositol 1,3,4-trisphosphate
1D-myo-inositol 1,3,4-trisphosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.3.1.3.86
- phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4,5-trisphosphate) + H2O = a 1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4- bisphosphate) + phosphate
|
 |
 |
 |
 |
 |
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4,5-trisphosphate)
|
+
|
H2O
|
=
|
1,2-diacyl-sn-glycero-3-phospho-(1D-myo-inositol-3,4- bisphosphate)
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

