 |
PDBsum entry 5onc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
5onc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.16
- acetyl-CoA C-acyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an acyl-CoA + acetyl-CoA = a 3-oxoacyl-CoA + CoA
|
 |
 |
 |
 |
 |
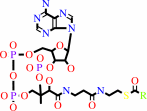 acyl-CoA
acyl-CoA
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
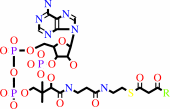 3-oxoacyl-CoA
3-oxoacyl-CoA
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
ACS Infect Dis
3:666-675
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis Is Controlled by a Redox-Sensitive Thiol Switch.
|
|
R.Lu,
C.M.Schaefer,
N.M.Nesbitt,
J.Kuper,
C.Kisker,
N.S.Sampson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB), is a
highly successful human pathogen and has infected approximately one-third of the
world's population. Multiple drug resistant (MDR) and extensively drug resistant
(XDR) TB strains and coinfection with HIV have increased the challenges of
successfully treating this disease pandemic. The metabolism of host cholesterol
by Mtb is an important factor for both its virulence and pathogenesis. In Mtb,
the cholesterol side chain is degraded through multiple cycles of β-oxidation
and FadA5 (Rv3546) catalyzes side chain thiolysis in the first two cycles.
Moreover, FadA5 is important during the chronic stage of infection in a mouse
model of Mtb infection. Here, we report the redox control of FadA5 catalytic
activity that results from reversible disulfide bond formation between
Cys59-Cys91 and Cys93-Cys377. Cys93 is the thiolytic nucleophile, and Cys377 is
the general acid catalyst for cleavage of the β-keto-acyl-CoA substrate. The
disulfide bond formed between the two catalytic residues Cys93 and Cys377 blocks
catalysis. The formation of the disulfide bonds is accompanied by a large domain
swap at the FadA5 dimer interface that serves to bring Cys93 and Cys377 in close
proximity for disulfide bond formation. The catalytic activity of FadA5 has a
midpoint potential of -220 mV, which is close to the Mtb mycothiol potential in
the activated macrophage. The redox profile of FadA5 suggests that FadA5 is
fully active when Mtb resides in the unactivated macrophage to maximize flux
into cholesterol catabolism. Upon activation of the macrophage, the oxidative
shift in the mycothiol potential will decrease the thiolytic activity by 50%.
Thus, the FadA5 midpoint potential is poised to rapidly restrict cholesterol
side chain degradation in response to oxidative stress from the host macrophage
environment.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

