 |
PDBsum entry 5ohc
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.2.1.208
- glucosylglycerate hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R)-2-O-(alpha-D-glucopyranosyl)-glycerate + H2O = (R)-glycerate + D-glucose
|
 |
 |
 |
 |
 |
(2R)-2-O-(alpha-D-glucopyranosyl)-glycerate
|
+
|
H2O
|
=
|
(R)-glycerate
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
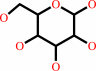 D-glucose
D-glucose
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
IUCrJ
6:572-585
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural characterization of a glucosylglycerate hydrolase provides insights into the molecular mechanism of mycobacterial recovery from nitrogen starvation.
|
|
T.B.Cereija,
S.Alarico,
E.C.Lourenço,
J.A.Manso,
M.R.Ventura,
N.Empadinhas,
S.Macedo-Ribeiro,
P.J.B.Pereira.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Bacteria are challenged to adapt to environmental variations in order to
survive. Under nutritional stress, several bacteria are able to slow down their
metabolism into a nonreplicating state and wait for favourable conditions. It is
almost universal that bacteria accumulate carbon stores to survive during this
nonreplicating state and to fuel rapid proliferation when the growth-limiting
stress disappears. Mycobacteria are exceedingly successful in their ability to
become dormant under harsh circumstances and to be able to resume growth when
conditions are favourable. Rapidly growing mycobacteria accumulate
glucosylglycerate under nitrogen-limiting conditions and quickly mobilize it
when nitrogen availability is restored. The depletion of intracellular
glucosyl-glycerate levels in Mycolicibacterium hassiacum (basonym
Mycobacterium hassiacum) was associated with the up-regulation of the
gene coding for glucosylglycerate hydrolase (GgH), an enzyme that is able to
hydrolyse glucosylglycerate to glycerate and glucose, a source of readily
available energy. Highly conserved among unrelated phyla, GgH is likely to be
involved in bacterial reactivation following nitrogen starvation, which in
addition to other factors driving mycobacterial recovery may also provide an
opportunity for therapeutic intervention, especially in the serious infections
caused by some emerging opportunistic pathogens of this group, such as
Mycobacteroides abscessus (basonym Mycobacterium abscessus). Using
a combination of biochemical methods and hybrid structural approaches, the
oligomeric organization of M. hassiacum GgH was determined and molecular
determinants of its substrate binding and specificity were unveiled.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

