 |
PDBsum entry 5hg0
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of pantoate-beta-alanine ligase from francisella tularensis complex with sam
|
|
Structure:
|
 |
Pantothenate synthetase. Chain: a, b. Synonym: ps,pantoate--beta-alanine ligase,pantoate-activating enzyme. Engineered: yes. Other_details: n-terminal cloning artifact sna
|
|
Source:
|
 |
Francisella tularensis subsp. Tularensis (strain schu s4 / schu 4). Organism_taxid: 177416. Strain: schu s4 / schu 4. Gene: panc, ftt_1390. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.187
|
R-free:
|
0.235
|
|
|
Authors:
|
 |
C.Chang,N.Maltseva,Y.Kim,L.Papazisi,W.F.Anderson,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
C.Chang
et al.
Crystal structure of pantoate-Beta-Alanine ligase fro francisella tularensis comlex with sam.
To be published,
.
|
 |
|
Date:
|
 |
|
07-Jan-16
|
Release date:
|
20-Jan-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5NF57
(PANC_FRATT) -
Pantothenate synthetase from Francisella tularensis subsp. tularensis (strain SCHU S4 / Schu 4)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
261 a.a.
261 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.2.1
- pantoate--beta-alanine ligase (AMP-forming).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Coenzyme A Biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(R)-pantoate + beta-alanine + ATP = (R)-pantothenate + AMP + diphosphate + H+
|
 |
 |
 |
 |
 |
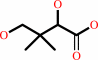 (R)-pantoate
(R)-pantoate
|
+
|
 beta-alanine
beta-alanine
|
+
|
 ATP
ATP
|
=
|
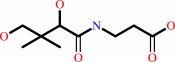 (R)-pantothenate
(R)-pantothenate
|
+
|
AMP
Bound ligand (Het Group name = )
matches with 56.25% similarity
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

