 |
PDBsum entry 5eiy

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Membrane protein
|
PDB id
|
|

|

|
5eiy
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.2.4.1.12
- cellulose synthase (UDP-forming).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
[(1->4)-beta-D-glucosyl](n) + UDP-alpha-D-glucose = [(1->4)-beta-D- glucosyl](n+1) + UDP + H+
|
 |
 |
 |
 |
 |
[(1->4)-beta-D-glucosyl](n)
|
+
|
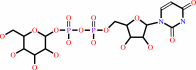 UDP-alpha-D-glucose
UDP-alpha-D-glucose
|
=
|
[(1->4)-beta-D- glucosyl](n+1)
|
+
|
 UDP
UDP
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 64.86% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
531:329-334
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Observing cellulose biosynthesis and membrane translocation in crystallo.
|
|
J.L.Morgan,
J.T.McNamara,
M.Fischer,
J.Rich,
H.M.Chen,
S.G.Withers,
J.Zimmer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Many biopolymers, including polysaccharides, must be translocated across at
least one membrane to reach their site of biological function. Cellulose is a
linear glucose polymer synthesized and secreted by a membrane-integrated
cellulose synthase. Here, in crystallo enzymology with the catalytically active
bacterial cellulose synthase BcsA-BcsB complex reveals structural snapshots of a
complete cellulose biosynthesis cycle, from substrate binding to polymer
translocation. Substrate- and product-bound structures of BcsA provide the basis
for substrate recognition and demonstrate the stepwise elongation of cellulose.
Furthermore, the structural snapshots show that BcsA translocates cellulose via
a ratcheting mechanism involving a 'finger helix' that contacts the polymer's
terminal glucose. Cooperating with BcsA's gating loop, the finger helix moves
'up' and 'down' in response to substrate binding and polymer elongation,
respectively, thereby pushing the elongated polymer into BcsA's transmembrane
channel. This mechanism is validated experimentally by tethering BcsA's finger
helix, which inhibits polymer translocation but not elongation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

