 |
PDBsum entry 5e8f
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structure of fully modified geranylgeranylated pde6c peptide in complex with pde6d
|
|
Structure:
|
 |
Retinal rod rhodopsin-sensitive cgmp 3',5'-cyclic phosphodiesterase subunit delta. Chain: a, c. Fragment: unp residues 2-150. Synonym: gmp-pde delta,protein p17. Engineered: yes. Cone cgmp-specific 3',5'-cyclic phosphodiesterase subunit alpha'. Chain: d, e.
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pde6d, pded. Expressed in: escherichia coli. Expression_system_taxid: 562. Gene: pde6c, pdea2. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.10Å
|
R-factor:
|
0.201
|
R-free:
|
0.256
|
|
|
Authors:
|
 |
E.K.Fansa,N.J.O'Reilly,S.A.Ismail,A.Wittinghofer
|
|
Key ref:
|
 |
E.K.Fansa
et al.
(2015).
The N- and C-terminal ends of RPGR can bind to PDE6δ.
Embo Rep,
16,
1583-1585.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
14-Oct-15
|
Release date:
|
18-Nov-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O43924
(PDE6D_HUMAN) -
Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
150 a.a.
147 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.4.35
- 3',5'-cyclic-GMP phosphodiesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3',5'-cyclic GMP + H2O = GMP + H+
|
 |
 |
 |
 |
 |
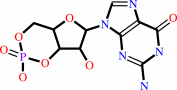 3',5'-cyclic GMP
3',5'-cyclic GMP
|
+
|
H2O
|
=
|
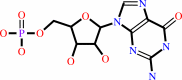 GMP
GMP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Embo Rep
16:1583-1585
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
The N- and C-terminal ends of RPGR can bind to PDE6δ.
|
|
E.K.Fansa,
N.J.O'Reilly,
S.Ismail,
A.Wittinghofer.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

