 |
PDBsum entry 5dnx
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of igpd from pyrococcus furiosus in complex with (r)-c348
|
|
Structure:
|
 |
Imidazoleglycerol-phosphate dehydratase. Chain: a, c, b. Synonym: igpd. Engineered: yes
|
|
Source:
|
 |
Pyrococcus furiosus. Organism_taxid: 2261. Gene: hisb, pf1660. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.132
|
R-free:
|
0.158
|
|
|
Authors:
|
 |
C.Bisson,K.L.Britton,S.E.Sedelnikova,H.F.Rodgers,T.C.Eadsforth, R.C.Viner,T.R.Hawkes,P.J.Baker,D.W.Rice
|
|
Key ref:
|
 |
C.Bisson
et al.
(2016).
Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew Chem Int Ed Engl,
55,
13485-13489.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
10-Sep-15
|
Release date:
|
05-Oct-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P58880
(HIS7_PYRFU) -
Imidazoleglycerol-phosphate dehydratase from Pyrococcus furiosus (strain ATCC 43587 / DSM 3638 / JCM 8422 / Vc1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
176 a.a.
176 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.19
- imidazoleglycerol-phosphate dehydratase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Histidine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate = 3-(imidazol-4-yl)-2- oxopropyl phosphate + H2O
|
 |
 |
 |
 |
 |
D-erythro-1-(imidazol-4-yl)glycerol 3-phosphate
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
=
|
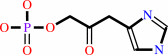 3-(imidazol-4-yl)-2- oxopropyl phosphate
3-(imidazol-4-yl)-2- oxopropyl phosphate
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Angew Chem Int Ed Engl
55:13485-13489
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
|
|
C.Bisson,
K.L.Britton,
S.E.Sedelnikova,
H.F.Rodgers,
T.C.Eadsforth,
R.C.Viner,
T.R.Hawkes,
P.J.Baker,
D.W.Rice.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

