 |
PDBsum entry 5cnp
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
X-ray crystal structure of spermidine n1-acetyltransferase from vibrio cholerae.
|
|
Structure:
|
 |
Spermidine n(1)-acetyltransferase. Chain: a, b, c, d, e, f. Synonym: sat,spermidine/spermine n(1)-acetyltransferase,ssat. Engineered: yes. Other_details: sna - cloning artifacts
|
|
Source:
|
 |
Vibrio cholerae serotype o1 (strain atcc 39315 / el tor inaba n16961). Organism_taxid: 243277. Strain: atcc 39315 / el tor inaba n16961. Gene: speg, vc_a0947. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.38Å
|
R-factor:
|
0.188
|
R-free:
|
0.236
|
|
|
Authors:
|
 |
J.Osipiuk,L.Volkart,S.Moy,W.F.Anderson,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
E.V.Filippova
et al.
(2015).
Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J Mol Biol,
427,
3538-3553.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
17-Jul-15
|
Release date:
|
29-Jul-15
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9KL03
(ATDA_VIBCH) -
Spermidine N(1)-acetyltransferase from Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
173 a.a.
170 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.57
- diamine N-acetyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an alkane-alpha,omega-diamine + acetyl-CoA = an N-acetylalkane- alpha,omega-diamine + CoA + H+
|
 |
 |
 |
 |
 |
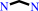 alkane-alpha,omega-diamine
alkane-alpha,omega-diamine
|
+
|
 acetyl-CoA
acetyl-CoA
|
=
|
N-acetylalkane- alpha,omega-diamine
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
427:3538-3553
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
|
|
E.V.Filippova,
S.Weigand,
J.Osipiuk,
O.Kiryukhina,
A.Joachimiak,
W.F.Anderson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The spermidine N-acetyltransferase SpeG is a dodecameric enzyme that catalyzes
the transfer of an acetyl group from acetyl coenzyme A to polyamines such as
spermidine and spermine. SpeG has an allosteric polyamine-binding site and
acetylating polyamines regulate their intracellular concentrations. The
structures of SpeG from Vibrio cholerae in complexes with polyamines and
cofactor have been characterized earlier. Here, we present the dodecameric
structure of SpeG from V. cholerae in a ligand-free form in three different
conformational states: open, intermediate and closed. All structures were
crystallized in C2 space group symmetry and contain six monomers in the
asymmetric unit cell. Two hexamers related by crystallographic 2-fold symmetry
form the SpeG dodecamer. The open and intermediate states have a unique open
dodecameric ring. This SpeG dodecamer is asymmetric except for the one 2-fold
axis and is unlike any known dodecameric structure. Using a fluorescence thermal
shift assay, size-exclusion chromatography with multi-angle light scattering,
small-angle X-ray scattering analysis, negative-stain electron microscopy and
structural analysis, we demonstrate that this unique open dodecameric state
exists in solution. Our combined results indicate that polyamines trigger
conformational changes and induce the symmetric closed dodecameric state of the
protein when they bind to their allosteric sites.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

