 |
PDBsum entry 5b4j

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5b4j
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.3.7.5
- phycocyanobilin:ferredoxin oxidoreductase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Biliverdin metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R,3Z)-phycocyanobilin + 4 oxidized [2Fe-2S]-[ferredoxin] = biliverdin IXalpha + 4 reduced [2Fe-2S]-[ferredoxin] + 4 H+
|
 |
 |
 |
 |
 |
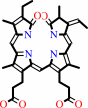 (3Z)-phycocyanobilin
(3Z)-phycocyanobilin
|
+
|
4
×
oxidized ferredoxin
|
=
|
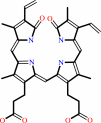 biliverdin IX-alpha
biliverdin IX-alpha
|
+
|
4
×
reduced ferredoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Febs Lett
590:3425-3434
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Atomic-resolution structure of the phycocyanobilin:ferredoxin oxidoreductase I86D mutant in complex with fully protonated biliverdin.
|
|
Y.Hagiwara,
K.Wada,
T.Irikawa,
H.Sato,
M.Unno,
K.Yamamoto,
K.Fukuyama,
M.Sugishima.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Phycocyanobilin:ferredoxin oxidoreductase (PcyA) catalyzes the reduction of
biliverdin (BV) to produce phycocyanobilin, a linear tetrapyrrole pigment used
for light harvesting and light sensing. Spectroscopic and HPLC analyses
inidicate that BV bound to the I86D mutant of PcyA is fully protonated (BVH(+) )
and can accept an electron, but I86D is unable to donate protons for the
reduction; therefore, compared to the wild-type PcyA, the I86D mutant stabilizes
BVH(+) . To elucidate the structural basis of the I86D mutation, we determined
the atomic-resolution structure of the I86D-BVH(+) complex and the protonation
states of the essential residues Asp105 and Glu76 in PcyA. Our study revealed
that Asp105 adopted a fixed conformation in the I86D mutant, although it had
dual conformations in wild-type PcyA which reflected the protonation states of
BV. Taken together with biochemical/spectroscopic results, our analysis of the
I86D-BVH(+) structure supports the hypothesis that flexibility of Asp105 is
essential for the catalytic activity of PcyA.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

