 |
PDBsum entry 5aih
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries. Ch55-sample-native
|
|
Structure:
|
 |
Limonene-1,2-epoxide hydrolase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Unidentified. Organism_taxid: 32644. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.42Å
|
R-factor:
|
0.153
|
R-free:
|
0.173
|
|
|
Authors:
|
 |
E.Ferrandi,C.Sayer,M.N.Isupov,C.Annovazzi,C.Marchesi,G.Iacobone, X.Peng,E.Bonch-Osmolovskaya,R.Wohlgemuth,J.A.Littlechild,D.Montia
|
|
Key ref:
|
 |
E.E.Ferrandi
et al.
(2015).
Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries.
Febs J,
282,
2879-2894.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
13-Feb-15
|
Release date:
|
17-Jun-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
No UniProt id for this chain
|
|

|

|
|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.3.2.8
- limonene-1,2-epoxide hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
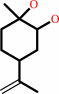
limonene 1,2-epoxide + H2O = limonene-1,2-diol
|
 |
 |
 |
 |
 |
 limonene 1,2-epoxide
limonene 1,2-epoxide
|
+
|
H2O
|
=
|
 limonene-1,2-diol
limonene-1,2-diol
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Febs J
282:2879-2894
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Discovery and characterization of thermophilic limonene-1,2-epoxide hydrolases from hot spring metagenomic libraries.
|
|
E.E.Ferrandi,
C.Sayer,
M.N.Isupov,
C.Annovazzi,
C.Marchesi,
G.Iacobone,
X.Peng,
E.Bonch-Osmolovskaya,
R.Wohlgemuth,
J.A.Littlechild,
D.Monti.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

