 |
PDBsum entry 5l2n

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/oxidoreductase inhibitor
|
PDB id
|
|

|

|
5l2n
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/oxidoreductase inhibitor
|
 |
|
Title:
|
 |
Structure of aldh1a1 in complex with buc25
|
|
Structure:
|
 |
Retinal dehydrogenase 1. Chain: a. Synonym: raldh1,aldh-e1,alhdii,aldehyde dehydrogenase family 1 member a1,aldehyde dehydrogenase,cytosolic. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: aldh1a1, aldc, aldh1, pumb1. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.216
|
R-free:
|
0.243
|
|
|
Authors:
|
 |
C.D.Buchman,T.D.Hurley
|
|
Key ref:
|
 |
C.D.Buchman
and
T.D.Hurley
(2017).
Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives.
J Med Chem,
60,
2439-2455.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-Aug-16
|
Release date:
|
08-Mar-17
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P00352
(AL1A1_HUMAN) -
Aldehyde dehydrogenase 1A1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
501 a.a.
494 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.2.1.19
- aminobutyraldehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-aminobutanal + NAD+ + H2O = 4-aminobutanoate + NADH + 2 H+
|
 |
 |
 |
 |
 |
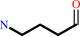 4-aminobutanal
4-aminobutanal
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
 4-aminobutanoate
4-aminobutanoate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.2.1.28
- benzaldehyde dehydrogenase (NAD(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
benzaldehyde + NAD+ + H2O = benzoate + NADH + 2 H+
|
 |
 |
 |
 |
 |
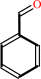 benzaldehyde
benzaldehyde
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
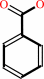 benzoate
benzoate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.2.1.3
- aldehyde dehydrogenase (NAD(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an aldehyde + NAD+ + H2O = a carboxylate + NADH + 2 H+
|
 |
 |
 |
 |
 |
 aldehyde
aldehyde
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
 carboxylate
carboxylate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.2.1.36
- retinal dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
retinal + NAD+ + H2O = retinoate + NADH + 2 H+
|
 |
 |
 |
 |
 |
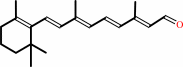 retinal
retinal
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
Bound ligand (Het Group name = )
matches with 60.71% similarity
|
=
|
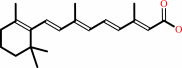 retinoate
retinoate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
 FAD
FAD
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Med Chem
60:2439-2455
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibition of the Aldehyde Dehydrogenase 1/2 Family by Psoralen and Coumarin Derivatives.
|
|
C.D.Buchman,
T.D.Hurley.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aldehyde dehydrogenase 2 (ALDH2), one of 19 ALDH superfamily members, catalyzes
the NAD(+)-dependent oxidation of aldehydes to their respective carboxylic
acids. In this study, we further characterized the inhibition of four psoralen
and coumarin derivatives toward ALDH2 and compared them to the ALDH2 inhibitor
daidzin for selectivity against five ALDH1/2 isoenzymes. Compound 2 (Ki = 19 nM)
binds within the aldehyde-binding site of the free enzyme species of ALDH2.
Thirty-three structural analogs were examined to develop a stronger SAR profile.
Seven compounds maintained or improved upon the selectivity toward one of the
five ALDH1/2 isoenzymes, including compound 36, a selective inhibitor for ALDH2
(Ki = 2.4 μM), and compound 32, which was 10-fold selective for ALDH1A1 (Ki =
1.2 μM) versus ALDH1A2. Further medicinal chemistry on the compounds' basic
scaffold could enhance the potency and selectivity for ALDH1A1 or ALDH2 and
generate chemical probes to examine the unique and overlapping functions of the
ALDH1/2 isoenzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

