 |
PDBsum entry 5dgs
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of human fpps in complex with the monophosphonate compound 15
|
|
Structure:
|
 |
Farnesyl pyrophosphate synthase. Chain: f. Fragment: unp residues 72-419. Synonym: fps,(2e,6e)-farnesyl diphosphate synthase, dimethylallyltranstransferase,farnesyl diphosphate synthase, geranyltranstransferase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: fdps, fps, kiaa1293. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008. Expression_system_variant: tuner.
|
|
Resolution:
|
 |
|
2.62Å
|
R-factor:
|
0.193
|
R-free:
|
0.243
|
|
|
Authors:
|
 |
J.M.Rondeau,E.Bourgier,S.Lehmann
|
|
Key ref:
|
 |
W.Jahnke
et al.
(2015).
A General Strategy for Targeting Drugs to Bone.
Angew Chem Int Ed Engl,
54,
14575-14579.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
28-Aug-15
|
Release date:
|
13-Jul-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14324
(FPPS_HUMAN) -
Farnesyl pyrophosphate synthase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
419 a.a.
343 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.1
- dimethylallyltranstransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Terpenoid biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + dimethylallyl diphosphate = (2E)- geranyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
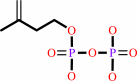 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
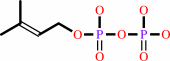 dimethylallyl diphosphate
dimethylallyl diphosphate
|
=
|
(2E)- geranyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.10
- (2E,6E)-farnesyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + (2E)-geranyl diphosphate = (2E,6E)-farnesyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
(2E)-geranyl diphosphate
|
=
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Angew Chem Int Ed Engl
54:14575-14579
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
A General Strategy for Targeting Drugs to Bone.
|
|
W.Jahnke,
G.Bold,
A.L.Marzinzik,
S.Ofner,
X.Pellé,
S.Cotesta,
E.Bourgier,
S.Lehmann,
C.Henry,
R.Hemmig,
F.Stauffer,
J.C.Hartwieg,
J.R.Green,
J.M.Rondeau.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Targeting drugs to their desired site of action can increase their safety and
efficacy. Bisphosphonates are prototypical examples of drugs targeted to bone.
However, bisphosphonate bone affinity is often considered too strong and cannot
be significantly modulated without losing activity on the enzymatic target,
farnesyl pyrophosphate synthase (FPPS). Furthermore, bisphosphonate bone
affinity comes at the expense of very low and variable oral bioavailability.
FPPS inhibitors were developed with a monophosphonate as a bone-affinity tag
that confers moderate affinity to bone, which can furthermore be tuned to the
desired level, and the relationship between structure and bone affinity was
evaluated by using an NMR-based bone-binding assay. The concept of targeting
drugs to bone with moderate affinity, while retaining oral bioavailability, has
broad application to a variety of other bone-targeted drugs.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

