 |
PDBsum entry 5cu7
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.1.3.62
- multiple inositol-polyphosphate phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
1D-myo-inositol hexakisphosphate + H2O = 1D-myo-inositol 1,2,4,5,6- pentakisphosphate + phosphate
|
|
2.
|
1D-myo-inositol 1,2,4,5,6-pentakisphosphate + H2O = 1D-myo-inositol 1,2,5,6-tetrakisphosphate + phosphate
|
|
3.
|
1D-myo-inositol 1,2,5,6-tetrakisphosphate + H2O = 1D-myo-inositol 1,2,6-trisphosphate + phosphate
|
|
4.
|
1D-myo-inositol 1,2,6-trisphosphate + H2O = 1D-myo-inositol 1,2- bisphosphate + phosphate
|
|
5.
|
1D-myo-inositol 1,2-bisphosphate + H2O = 1D-myo-inositol 2-phosphate + phosphate
|
|
 |
 |
 |
 |
 |
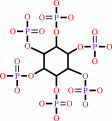 1D-myo-inositol hexakisphosphate
1D-myo-inositol hexakisphosphate
|
+
|
H2O
|
=
|
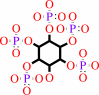 1D-myo-inositol 1,2,4,5,6- pentakisphosphate
1D-myo-inositol 1,2,4,5,6- pentakisphosphate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 1D-myo-inositol 1,2,4,5,6-pentakisphosphate
1D-myo-inositol 1,2,4,5,6-pentakisphosphate
|
+
|
H2O
|
=
|
1D-myo-inositol 1,2,5,6-tetrakisphosphate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
1D-myo-inositol 1,2,5,6-tetrakisphosphate
|
+
|
H2O
|
=
|
1D-myo-inositol 1,2,6-trisphosphate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
1D-myo-inositol 1,2,6-trisphosphate
|
+
|
H2O
|
=
|
1D-myo-inositol 1,2- bisphosphate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
1D-myo-inositol 1,2-bisphosphate
|
+
|
H2O
|
=
|
1D-myo-inositol 2-phosphate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.1.3.80
- 2,3-bisphosphoglycerate 3-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2R)-2,3-bisphosphoglycerate + H2O = (2R)-2-phosphoglycerate + phosphate
|
 |
 |
 |
 |
 |
(2R)-2,3-bisphosphoglycerate
|
+
|
H2O
|
=
|
(2R)-2-phosphoglycerate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

