 |
PDBsum entry 5amm

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
5amm
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of leishmania major peroxidase d211n mutant
|
|
Structure:
|
 |
Ascorbate peroxidase. Chain: a, b. Fragment: c-terminal catalytic domain, residues 35-303. Synonym: cytochromE C peroxidase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Leishmania major. Organism_taxid: 5664. Strain: friedlin. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.09Å
|
R-factor:
|
0.187
|
R-free:
|
0.245
|
|
|
Authors:
|
 |
G.Chreifi,J.B.Fields,S.A.Hollingsworth,M.Heyden,A.P.Arce,H.I.Magana- Garcia,T.L.Poulos,D.J.Tobias
|
|
Key ref:
|
 |
J.B.Fields
et al.
(2015).
"Bind and Crawl" Association Mechanism of Leishmania major Peroxidase and Cytochrome c Revealed by Brownian and Molecular Dynamics Simulations.
Biochemistry,
54,
7272-7282.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
11-Mar-15
|
Release date:
|
09-Dec-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q4Q3K2
(Q4Q3K2_LEIMA) -
Ascorbate peroxidase from Leishmania major
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
303 a.a.
267 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 2 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.11.1.11
- L-ascorbate peroxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ascorbate + H2O2 = L-dehydroascorbate + 2 H2O
|
 |
 |
 |
 |
 |
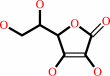 L-ascorbate
L-ascorbate
|
+
|
 H2O2
H2O2
|
=
|
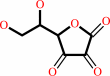 L-dehydroascorbate
L-dehydroascorbate
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
54:7272-7282
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
"Bind and Crawl" Association Mechanism of Leishmania major Peroxidase and Cytochrome c Revealed by Brownian and Molecular Dynamics Simulations.
|
|
J.B.Fields,
S.A.Hollingsworth,
G.Chreifi,
M.Heyden,
A.P.Arce,
H.I.Magaña-Garcia,
T.L.Poulos,
D.J.Tobias.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Leishmania major, the parasitic causative agent of leishmaniasis, produces a
heme peroxidase (LmP), which catalyzes the peroxidation of mitochondrial
cytochrome c (LmCytc) for protection from reactive oxygen species produced by
the host. The association of LmP and LmCytc, which is known from kinetics
measurements to be very fast (∼10(8) M(-1) s(-1)), does not involve major
conformational changes and has been suggested to be dominated by electrostatic
interactions. We used Brownian dynamics simulations to investigate the mechanism
of formation of the LmP-LmCytc complex. Our simulations confirm the importance
of electrostatic interactions involving the negatively charged D211 residue at
the LmP active site, and reveal a previously unrecognized role in complex
formation for negatively charged residues in helix A of LmP. The crystal
structure of the D211N mutant of LmP reported herein is essentially identical to
that of wild-type LmP, reinforcing the notion that it is the loss of charge at
the active site, and not a change in structure, that reduces the association
rate of the D211N variant of LmP. The Brownian dynamics simulations further show
that complex formation occurs via a "bind and crawl" mechanism, in
which LmCytc first docks to a location on helix A that is far from the active
site, forming an initial encounter complex, and then moves along helix A to the
active site. An atomistic molecular dynamics simulation confirms the helix A
binding site, and steady state activity assays and stopped-flow kinetics
measurements confirm the role of helix A charges in the association mechanism.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

