 |
PDBsum entry 4zwl

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4zwl
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
2.60 angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betb) h448f/y450l double mutant from staphylococcus aureus in complex with NAD+ and bme-free cys289
|
|
Structure:
|
 |
Betaine-aldehyde dehydrogenase. Chain: a, b, c, d, e, f, g, h. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Staphylococcus aureus (strain col). Organism_taxid: 93062. Strain: col. Gene: betb, sacol2628. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.60Å
|
R-factor:
|
0.162
|
R-free:
|
0.232
|
|
|
Authors:
|
 |
A.S.Halavaty,G.Minasov,C.Chen,J.C.Joo,A.F.Yakunin,W.F.Anderson,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
A.S.Halavaty
et al.
2.60 angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betb) h448f/y450l double muta staphylococcus aureus in complex with NAD+ and bme-Fr cys289.
To be published,
.
|
 |
|
Date:
|
 |
|
19-May-15
|
Release date:
|
27-May-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0A0M3KL40
(A0A0M3KL40_STAAC) -
Betaine-aldehyde dehydrogenase from Staphylococcus aureus (strain COL)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
496 a.a.
499 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.2.1.8
- betaine-aldehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
betaine aldehyde + NAD+ + H2O = glycine betaine + NADH + 2 H+
|
 |
 |
 |
 |
 |
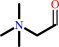 betaine aldehyde
betaine aldehyde
|
+
|
 NAD(+)
NAD(+)
|
+
|
H2O
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
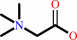 glycine betaine
glycine betaine
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

