 |
PDBsum entry 4zv4
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Translation
|
 |
|
Title:
|
 |
Structure of tse6 in complex with ef-tu
|
|
Structure:
|
 |
Elongation factor tu. Chain: a, b. Synonym: ef-tu. Engineered: yes. Tse6. Chain: c, d. Fragment: unp residues 265-430. Engineered: yes
|
|
Source:
|
 |
Pseudomonas aeruginosa (strain atcc 15692 / pao1 / 1c / prs 101 / lmg 12228). Organism_taxid: 208964. Strain: atcc 15692 / pao1 / 1c / prs 101 / lmg 12228. Gene: tufa, pa4265, tufb, pa4277. Expressed in: escherichia coli. Expression_system_taxid: 511693. Pseudomonas aeruginosa. Gene: pa0093.
|
|
Resolution:
|
 |
|
3.50Å
|
R-factor:
|
0.233
|
R-free:
|
0.283
|
|
|
Authors:
|
 |
J.C.Whitney,S.Sawai,H.Robinson,J.D.Mougous
|
|
Key ref:
|
 |
J.C.Whitney
et al.
(2015).
An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells.
Cell,
163,
607-619.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
18-May-15
|
Release date:
|
11-Nov-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B:
E.C.3.6.5.3
- protein-synthesizing GTPase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
GTP + H2O = GDP + phosphate + H+
|
 |
 |
 |
 |
 |
 GTP
GTP
|
+
|
H2O
|
=
|
GDP
Bound ligand (Het Group name = )
matches with 72.97% similarity
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains C, D:
E.C.3.2.2.5
- NAD(+) glycohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
NAD+ + H2O = ADP-D-ribose + nicotinamide + H+
|
 |
 |
 |
 |
 |
 NAD(+)
NAD(+)
|
+
|
H2O
|
=
|
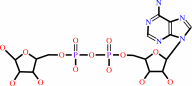 ADP-D-ribose
ADP-D-ribose
|
+
|
 nicotinamide
nicotinamide
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Cell
163:607-619
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
An interbacterial NAD(P)(+) glycohydrolase toxin requires elongation factor Tu for delivery to target cells.
|
|
J.C.Whitney,
D.Quentin,
S.Sawai,
M.LeRoux,
B.N.Harding,
H.E.Ledvina,
B.Q.Tran,
H.Robinson,
Y.A.Goo,
D.R.Goodlett,
S.Raunser,
J.D.Mougous.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Type VI secretion (T6S) influences the composition of microbial communities by
catalyzing the delivery of toxins between adjacent bacterial cells. Here, we
demonstrate that a T6S integral membrane toxin from Pseudomonas aeruginosa,
Tse6, acts on target cells by degrading the universally essential dinucleotides
NAD(+) and NADP(+). Structural analyses of Tse6 show that it resembles
mono-ADP-ribosyltransferase proteins, such as diphtheria toxin, with the
exception of a unique loop that both excludes proteinaceous ADP-ribose acceptors
and contributes to hydrolysis. We find that entry of Tse6 into target cells
requires its binding to an essential housekeeping protein, translation
elongation factor Tu (EF-Tu). These proteins participate in a larger assembly
that additionally directs toxin export and provides chaperone activity.
Visualization of this complex by electron microscopy defines the architecture of
a toxin-loaded T6S apparatus and provides mechanistic insight into intercellular
membrane protein delivery between bacteria.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

