 |
PDBsum entry 4zv3
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of the n- and c-terminal domains of mouse acyl-coa thioesterase 7
|
|
Structure:
|
 |
Cytosolic acyl coenzyme a thioester hydrolase. Chain: a, b, c. Fragment: unp residues 55-369. Synonym: acyl-coa thioesterase 7,brain acyl-coa hydrolase,bach,cte- iia,cte-ii,long chain acyl-coa thioester hydrolase. Engineered: yes
|
|
Source:
|
 |
Mus musculus. Mouse. Organism_taxid: 10090. Gene: acot7, bach. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
3.10Å
|
R-factor:
|
0.244
|
R-free:
|
0.296
|
|
|
Authors:
|
 |
C.M.D.Swarbrick,J.K.Forwood
|
|
Key ref:
|
 |
C.M.D.Swarbrick
and
j.k.forwood
Crystal structure of the n- And c-Terminal domains of acyl-Coa thioesterase 7.
To be published,
.
|
 |
|
Date:
|
 |
|
18-May-15
|
Release date:
|
03-Jun-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q91V12
(BACH_MOUSE) -
Cytosolic acyl coenzyme A thioester hydrolase from Mus musculus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
381 a.a.
299 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.2.2
- palmitoyl-CoA hydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hexadecanoyl-CoA + H2O = hexadecanoate + CoA + H+
|
 |
 |
 |
 |
 |
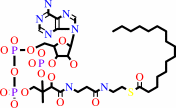 hexadecanoyl-CoA
hexadecanoyl-CoA
|
+
|
H2O
|
=
|
hexadecanoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 CoA
CoA
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

