 |
PDBsum entry 4zt2

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Ligase/ligase inhibitor
|
PDB id
|
|

|

|
4zt2
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.10
- methionine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Met) + L-methionine + ATP = L-methionyl-tRNA(Met) + AMP + diphosphate
|
 |
 |
 |
 |
 |
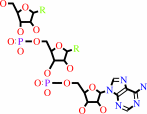 tRNA(Met)
tRNA(Met)
|
+
|
L-methionine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
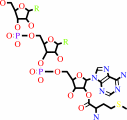 L-methionyl-tRNA(Met)
L-methionyl-tRNA(Met)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acs Infect Dis
2:399-404
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
5-Fluoroimidazo[4,5-b]pyridine Is a Privileged Fragment That Conveys Bioavailability to Potent Trypanosomal Methionyl-tRNA Synthetase Inhibitors.
|
|
Z.Zhang,
C.Y.Koh,
R.M.Ranade,
S.Shibata,
J.R.Gillespie,
M.A.Hulverson,
W.Huang,
J.Nguyen,
N.Pendem,
M.H.Gelb,
C.L.Verlinde,
W.G.Hol,
F.S.Buckner,
E.Fan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Fluorination is a well-known strategy for improving the bioavailability of drug
molecules. However, its impact on efficacy is not easily predicted. On the basis
of inhibitor-bound protein crystal structures, we found a beneficial
fluorination spot for inhibitors targeting methionyl-tRNA synthetase of
Trypanosoma brucei. In particular, incorporating 5-fluoroimidazo[4,5-b]pyridine
into inhibitors leads to central nervous system bioavailability and maintained
or even improved efficacy.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

