 |
PDBsum entry 4zsw

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
4zsw
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase/transferase inhibitor
|
 |
|
Title:
|
 |
Pig brain gaba-at inactivated by (e)-(1s,3s)-3-amino-4- fluoromethylenyl-1-cyclopentanoic acid
|
|
Structure:
|
 |
4-aminobutyrate aminotransferase, mitochondrial. Chain: a, b, c, d. Fragment: unp residues 39-499. Synonym: (s)-3-amino-2-methylpropionate transaminase,gaba aminotransferase,gaba-at,gamma-amino-n-butyrate transaminase,gaba-t, l-aibat. Engineered: yes
|
|
Source:
|
 |
Sus scrofa. Pig. Organism_taxid: 9823. Gene: abat, gabat. Expressed in: sus scrofa domesticus. Expression_system_taxid: 9825
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.159
|
R-free:
|
0.195
|
|
|
Authors:
|
 |
R.Wu,H.Lee,H.V.Le,E.Doud,R.Sanishvili,P.Compton,N.L.Kelleher, R.B.Silverman,D.Liu
|
|
Key ref:
|
 |
H.Lee
et al.
(2015).
Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
Acs Chem Biol,
10,
2087-2098.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
14-May-15
|
Release date:
|
08-Jul-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P80147
(GABT_PIG) -
4-aminobutyrate aminotransferase, mitochondrial from Sus scrofa
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
500 a.a.
461 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.6.1.19
- 4-aminobutyrate--2-oxoglutarate transaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-aminobutanoate + 2-oxoglutarate = succinate semialdehyde + L-glutamate
|
 |
 |
 |
 |
 |
4-aminobutanoate
Bound ligand (Het Group name = )
matches with 45.45% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
succinate semialdehyde
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
+
|
 L-glutamate
L-glutamate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
RW2)
matches with 55.56% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.2.6.1.22
- (S)-3-amino-2-methylpropionate transaminase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-3-amino-2-methylpropanoate + 2-oxoglutarate = 2-methyl-3- oxopropanoate + L-glutamate
|
 |
 |
 |
 |
 |
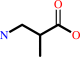 (S)-3-amino-2-methylpropanoate
(S)-3-amino-2-methylpropanoate
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
=
|
2-methyl-3- oxopropanoate
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
L-glutamate
Bound ligand (Het Group name = )
matches with 40.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acs Chem Biol
10:2087-2098
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Mechanism of Inactivation of GABA Aminotransferase by (E)- and (Z)-(1S,3S)-3-Amino-4-fluoromethylenyl-1-cyclopentanoic Acid.
|
|
H.Lee,
H.V.Le,
R.Wu,
E.Doud,
R.Sanishvili,
J.F.Kellie,
P.D.Compton,
B.Pachaiyappan,
D.Liu,
N.L.Kelleher,
R.B.Silverman.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

