 |
PDBsum entry 4znd
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
2.55 angstrom resolution structure of 3-phosphoshikimate 1- carboxyvinyltransferase (aroa) from coxiella burnetii in complex with shikimate-3-phosphate, phosphate, and potassium
|
|
Structure:
|
 |
3-phosphoshikimate 1-carboxyvinyltransferase. Chain: a. Synonym: 5-enolpyruvylshikimate-3-phosphate synthase,epsps. Engineered: yes
|
|
Source:
|
 |
Coxiella burnetii (strain rsa 493 / nine mile phase i). Organism_taxid: 227377. Strain: rsa 493 / nine mile phase i. Gene: aroa, cbu_0526. Expressed in: escherichia coli. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
2.55Å
|
R-factor:
|
0.169
|
R-free:
|
0.238
|
|
|
Authors:
|
 |
S.H.Light,G.Minasov,S.N.Krishna,K.Kwon,W.F.Anderson,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
S.H.Light
et al.
2.55 angstrom resolution structure of 3-Phosphoshikim 1-Carboxyvinyltransferase (aroa) from coxiella burnet complex with shikimate-3-Phosphate, Phosphate, And po.
To be published,
.
|
 |
|
Date:
|
 |
|
04-May-15
|
Release date:
|
13-May-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q83E11
(AROA_COXBU) -
3-phosphoshikimate 1-carboxyvinyltransferase from Coxiella burnetii (strain RSA 493 / Nine Mile phase I)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
438 a.a.
436 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.19
- 3-phosphoshikimate 1-carboxyvinyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Shikimate and Chorismate Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-phosphoshikimate + phosphoenolpyruvate = 5-O-(1-carboxyvinyl)-3- phosphoshikimate + phosphate
|
 |
 |
 |
 |
 |
3-phosphoshikimate
|
+
|
phosphoenolpyruvate
Bound ligand (Het Group name = )
matches with 52.94% similarity
|
=
|
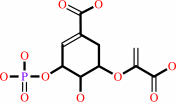 5-O-(1-carboxyvinyl)-3- phosphoshikimate
5-O-(1-carboxyvinyl)-3- phosphoshikimate
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

