 |
PDBsum entry 4zm9
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of circularly permuted human asparaginase-like protein 1
|
|
Structure:
|
 |
Isoaspartyl peptidase/l-asparaginase. Chain: a, b, c, d. Synonym: asparaginase-like protein 1,beta-aspartyl-peptidase, isoaspartyl dipeptidase,l-asparagine amidohydrolase. Engineered: yes. Other_details: circular permutation
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: asrgl1, alp, crash. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.51Å
|
R-factor:
|
0.206
|
R-free:
|
0.233
|
|
|
Authors:
|
 |
W.Z.Li,Y.Zhang
|
|
Key ref:
|
 |
W.Li
et al.
(2016).
Intramolecular Cleavage of the hASRGL1 Homodimer Occurs in Two Stages.
Biochemistry,
55,
960-969.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
02-May-15
|
Release date:
|
17-Feb-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q7L266
(ASGL1_HUMAN) -
Isoaspartyl peptidase/L-asparaginase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
308 a.a.
296 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.3.4.19.5
- beta-aspartyl-peptidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Cleavage of a beta-linked aspartic residue from the N-terminus of a polypeptide.
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.1.1
- asparaginase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-asparagine + H2O = L-aspartate + NH4+
|
 |
 |
 |
 |
 |
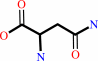 L-asparagine
L-asparagine
|
+
|
H2O
|
=
|
L-aspartate
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
55:960-969
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Intramolecular Cleavage of the hASRGL1 Homodimer Occurs in Two Stages.
|
|
W.Li,
S.Irani,
A.Crutchfield,
K.Hodge,
W.Matthews,
P.Patel,
Y.J.Zhang,
E.Stone.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The human asparaginase-like protein 1 (hASRGL1) is a member of the N-terminal
nucleophile (Ntn) family that hydrolyzes l-asparagine and
isoaspartyl-dipeptides. The nascent protein folds into an αβ-βα sandwich
fold homodimer that cleaves its own peptide backbone at the G167-T168 bond,
resulting in the active form of the enzyme. However, biophysical studies of
hASRGL1 are difficult because of the curious fact that intramolecular cleavage
of the G167-T168 peptide bond reaches only ≤50% completion. We capitalized
upon our previous observation that intramolecular processing increases
thermostability and developed a differential scanning fluorimetry assay that
allowed direct detection of distinct processing intermediates for the first
time. A kinetic analysis of these intermediates revealed that cleavage of one
subunit of the hASRGL1 subunit drastically reduces the processing rate of the
adjacent monomer, and a mutagenesis study showed that stabilization of the dimer
interface plays a critical role in this process. We also report a comprehensive
analysis of conserved active site residues and delineate their relative roles in
autoprocessing and substrate hydrolysis. In addition to glycine, which was
previously reported to selectively accelerate hASRGL1 cleavage, we identified
several novel small molecule activators that also promote intramolecular
processing. The structure-activity analysis supports the hypothesis that
multiple negatively charged small molecules interact within the active site of
hASRGL1 to act as a base in promoting cleavage. Overall, our investigation
provides a mechanistic understanding of the maturation process of this Ntn
hydrolase family member.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

