 |
PDBsum entry 4zhs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4zhs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.2.1.11
- aspartate-semialdehyde dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Lysine biosynthesis (early stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-aspartate 4-semialdehyde + phosphate + NADP+ = 4-phospho-L-aspartate + NADPH + H+
|
 |
 |
 |
 |
 |
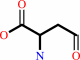 L-aspartate 4-semialdehyde
L-aspartate 4-semialdehyde
|
+
|
 phosphate
phosphate
|
+
|
 NADP(+)
NADP(+)
|
=
|
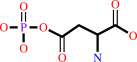 4-phospho-L-aspartate
4-phospho-L-aspartate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Sci Rep
6:21067
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural Insights into the Tetrameric State of Aspartate-β-semialdehyde Dehydrogenases from Fungal Species.
|
|
Q.Li,
Z.Mu,
R.Zhao,
G.Dahal,
R.E.Viola,
T.Liu,
Q.Jin,
S.Cui.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aspartate-β-semialdehyde dehydrogenase (ASADH) catalyzes the second reaction in
the aspartate pathway, a pathway required for the biosynthesis of one fifth of
the essential amino acids in plants and microorganisms. Microarray analysis of a
fungal pathogen T. rubrum responsible for most human dermatophytoses identified
the upregulation of ASADH (trASADH) expression when the fungus is exposed to
human skin, underscoring its potential as a drug target. Here we report the
crystal structure of trASADH, revealing a tetrameric ASADH with a GAPDH-like
fold. The tetramerization of trASADH was confirmed by sedimentation and SAXS
experiments. Native PAGE demonstrated that this ASADH tetramerization is
apparently universal in fungal species, unlike the functional dimer that is
observed in all bacterial ASADHs. The helical subdomain in dimeric bacteria
ASADH is replaced by the cover loop in archaeal/fungal ASADHs, presenting the
determinant for this altered oligomerization. Mutations that disrupt the
tetramerization of trASADH also abolish the catalytic activity, suggesting that
the tetrameric state is required to produce the active fungal enzyme form. Our
findings provide a basis to categorize ASADHs into dimeric and tetrameric
enzymes, adopting a different orientation for NADP binding and offer a
structural framework for designing drugs that can specifically target the fungal
pathogens.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

