 |
PDBsum entry 4zgy

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Lyase/lyase inhibitor
|
PDB id
|
|

|

|
4zgy
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chain A:
E.C.4.1.1.17
- ornithine decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Spermine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-ornithine + H+ = putrescine + CO2
|
 |
 |
 |
 |
 |
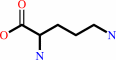 L-ornithine
L-ornithine
|
+
|
H(+)
|
=
|
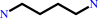 putrescine
putrescine
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
112:11229-11234
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis of antizyme-mediated regulation of polyamine homeostasis.
|
|
H.Y.Wu,
S.F.Chen,
J.Y.Hsieh,
F.Chou,
Y.H.Wang,
W.T.Lin,
P.Y.Lee,
Y.J.Yu,
L.Y.Lin,
T.S.Lin,
C.L.Lin,
G.Y.Liu,
S.R.Tzeng,
H.C.Hung,
N.L.Chan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Polyamines are organic polycations essential for cell growth and
differentiation; their aberrant accumulation is often associated with diseases,
including many types of cancer. To maintain polyamine homeostasis, the catalytic
activity and protein abundance of ornithine decarboxylase (ODC), the committed
enzyme for polyamine biosynthesis, are reciprocally controlled by the regulatory
proteins antizyme isoform 1 (Az1) and antizyme inhibitor (AzIN). Az1 suppresses
polyamine production by inhibiting the assembly of the functional ODC homodimer
and, most uniquely, by targeting ODC for ubiquitin-independent proteolytic
destruction by the 26S proteasome. In contrast, AzIN positively regulates
polyamine levels by competing with ODC for Az1 binding. The structural basis of
the Az1-mediated regulation of polyamine homeostasis has remained elusive. Here
we report crystal structures of human Az1 complexed with either ODC or AzIN.
Structural analysis revealed that Az1 sterically blocks ODC homodimerization.
Moreover, Az1 binding triggers ODC degradation by inducing the exposure of a
cryptic proteasome-interacting surface of ODC, which illustrates how a substrate
protein may be primed upon association with Az1 for ubiquitin-independent
proteasome recognition. Dynamic and functional analyses further indicated that
the Az1-induced binding and degradation of ODC by proteasome can be decoupled,
with the intrinsically disordered C-terminal tail fragment of ODC being required
only for degradation but not binding. Finally, the AzIN-Az1 structure suggests
how AzIN may effectively compete with ODC for Az1 to restore polyamine
production. Taken together, our findings offer structural insights into the
Az-mediated regulation of polyamine homeostasis and proteasomal degradation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

