 |
PDBsum entry 4zg5
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Structural and functional insights into survival endonuclease, an important virulence factor of brucella abortus
|
|
Structure:
|
 |
5'-nucleotidase sure. Chain: d, a, c, g. Synonym: survival endonuclease,nucleoside 5'-monophosphate phosphohydrolase. Engineered: yes
|
|
Source:
|
 |
Brucella abortus s19. Organism_taxid: 430066. Strain: s19. Gene: sure, babs19_i08450. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.200
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
K.F.Tarique,S.A.Abdul Rehman,S.Devi,S.Gourinath
|
|
Key ref:
|
 |
K.F.Tarique
et al.
(2016).
Structural and functional insights into the stationary-phase survival protein SurE, an important virulence factor of Brucella abortus.
Acta Crystallogr F Struct Biol Commun,
72,
386-396.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
22-Apr-15
|
Release date:
|
06-May-15
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
B2S5B9
(SURE_BRUA1) -
5'-nucleotidase SurE from Brucella abortus (strain S19)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
255 a.a.
252 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.5
- 5'-nucleotidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a ribonucleoside 5'-phosphate + H2O = a ribonucleoside + phosphate
|
 |
 |
 |
 |
 |
ribonucleoside 5'-phosphate
|
+
|
H2O
|
=
|
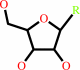 ribonucleoside
ribonucleoside
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr F Struct Biol Commun
72:386-396
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural and functional insights into the stationary-phase survival protein SurE, an important virulence factor of Brucella abortus.
|
|
K.F.Tarique,
S.A.Abdul Rehman,
S.Devi,
P.Tomar,
S.Gourinath.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The stationary-phase survival protein SurE from Brucella abortus (BaSurE) is a
metal-dependent phosphatase that is essential for the survival of this bacterium
in the stationary phase of its life cycle. Here, BaSurE has been biochemically
characterized and its crystal structure has been determined to a resolution of
1.9 Å. BaSurE was found to be a robust enzyme, showing activity over wide
ranges of temperature and pH and with various phosphoester substrates. The
active biomolecule is a tetramer and each monomer was found to consist of two
domains: an N-terminal domain, which forms an approximate α + β fold, and a
C-terminal domain that belongs to the α/β class. The active site lies at the
junction of these two domains and was identified by the presence of conserved
negatively charged residues and a bound Mg(2+) ion. Comparisons of BaSurE with
its homologues have revealed both common features and differences in this class
of enzymes. The number and arrangement of some of the equivalent secondary
structures, which are seen to differ between BaSurE and its homologues, are
responsible for a difference in the size of the active-site area and the overall
oligomeric state of this enzyme in other organisms. As it is absent in mammals,
it has the potential to be a drug target.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

