 |
PDBsum entry 4z82

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4z82
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.20
- cysteine dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-cysteine + O2 = 3-sulfino-L-alanine + H+
|
 |
 |
 |
 |
 |
L-cysteine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 O2
O2
|
=
|
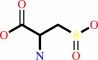 3-sulfino-L-alanine
3-sulfino-L-alanine
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+); NADH or NADPH
|
 |
 |
 |
 |
 |
Fe(2+)
|
 NADH
NADH
|
or
|
 NADPH
NADPH
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Inorg Chem
21:501-510
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Influence of cysteine 164 on active site structure in rat cysteine dioxygenase.
|
|
M.Fellner,
E.Siakkou,
A.S.Faponle,
E.P.Tchesnokov,
S.P.de Visser,
S.M.Wilbanks,
G.N.Jameson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cysteine dioxygenase is a non-heme mononuclear iron enzyme with unique
structural features, namely an intramolecular thioether cross-link between
cysteine 93 and tyrosine 157, and a disulfide bond between substrate L-cysteine
and cysteine 164 in the entrance channel to the active site. We investigated how
these posttranslational modifications affect catalysis through a kinetic,
crystallographic and computational study. The enzyme kinetics of a C164S variant
are identical to WT, indicating that disulfide formation at C164 does not
significantly impair access to the active site at physiological pH. However, at
high pH, the cysteine-tyrosine cross-link formation is enhanced in C164S. This
supports the view that disulfide formation at position 164 can limit access to
the active site. The C164S variant yielded crystal structures of unusual clarity
in both resting state and with cysteine bound. Both show that the iron in the
cysteine-bound complex is a mixture of penta- and hexa-coordinate with a water
molecule taking up the final site (60 % occupancy), which is where dioxygen is
believed to coordinate during turnover. The serine also displays stronger
hydrogen bond interactions to a water bound to the amine of the substrate
cysteine. However, the interactions between cysteine and iron appear unchanged.
DFT calculations support this and show that WT and C164S have similar binding
energies for the water molecule in the final site. This variant therefore
provides evidence that WT also exists in an equilibrium between penta- and
hexa-coordinate forms and the presence of the sixth ligand does not strongly
affect dioxygen binding.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

