 |
PDBsum entry 4z1b
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Structure of h204a mutant kdo8ps from h.Pylori
|
|
Structure:
|
 |
2-dehydro-3-deoxyphosphooctonate aldolase. Chain: a, b. Synonym: 3-deoxy-d-manno-octulosonic acid 8-phosphate synthase,kdo-8- phosphate synthase,kdops,phospho-2-dehydro-3-deoxyoctonate aldolase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Helicobacter pylori (strain atcc 700392 / 26695). Organism_taxid: 85962. Strain: atcc 700392 / 26695. Gene: kdsa, hp_0003. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.186
|
R-free:
|
0.238
|
|
|
Authors:
|
 |
B.J.Lee,S.Cho,H.Im,H.J.Yoon
|
|
Key ref:
|
 |
S.Cho
et al.
(2016).
Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
Eur J Med Chem,
108,
188-202.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
27-Mar-15
|
Release date:
|
09-Mar-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P56060
(KDSA_HELPY) -
2-dehydro-3-deoxyphosphooctonate aldolase from Helicobacter pylori (strain ATCC 700392 / 26695)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
276 a.a.
256 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.55
- 3-deoxy-8-phosphooctulonate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-arabinose 5-phosphate + phosphoenolpyruvate + H2O = 3-deoxy-alpha-D- manno-2-octulosonate-8-phosphate + phosphate
|
 |
 |
 |
 |
 |
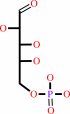 D-arabinose 5-phosphate
D-arabinose 5-phosphate
|
+
|
 phosphoenolpyruvate
phosphoenolpyruvate
|
+
|
H2O
|
=
|
3-deoxy-alpha-D- manno-2-octulosonate-8-phosphate
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Eur J Med Chem
108:188-202
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification of novel scaffolds for potential anti-Helicobacter pylori agents based on the crystal structure of H. pylori 3-deoxy-d-manno-octulosonate 8-phosphate synthase (HpKDO8PS).
|
|
S.Cho,
H.Im,
K.Y.Lee,
J.Chen,
H.J.Kang,
H.J.Yoon,
K.H.Min,
K.R.Lee,
H.J.Park,
B.J.Lee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structure of 3-deoxy-d-manno-octulosonate-8-phosphate synthase
(KDO8PS) from Helicobacter pylori (HpKDO8PS) was determined alone and within
various complexes, revealing an extra helix (HE) that is absent in the
structures of KDO8PS from other organisms. In contrast to the metal coordination
of the KDO8PS enzyme from Aquifex aeolicus, HpKDO8PS is specifically coordinated
with Cd(2+) or Zn(2+) ions, and isothermal titration calorimetry (ITC) and
differential scanning fluorimetry (DSF) revealed that Cd(2+) thermally
stabilizes the protein structure more efficiently than Zn(2+). In the
substrate-bound structure, water molecules play a key role in fixing residues in
the proper configuration to achieve a compact structure. Using the structures of
HpKDO8PS and API [arabinose 5-phosphate (A5P) and phosphoenolpyruvate (PEP)
bisubstrate inhibitor], we generated 21 compounds showing potential
HpKDO8PS-binding properties via in silico virtual screening. The capacity of
three, avicularin, hyperin, and MC181, to bind to HpKDO8PS was confirmed through
saturation transfer difference (STD) experiments, and we identified their
specific ligand binding modes by combining competition experiments and docking
simulation analysis. Hyperin was confirmed to bind to the A5P binding site,
primarily via hydrophilic interaction, whereas MC181 bound to both the PEP and
A5P binding sites through hydrophilic and hydrophobic interactions. These
results were consistent with the epitope mapping by STD. Our results are
expected to provide clues for the development of HpKDO8PS inhibitors.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

