 |
PDBsum entry 4xuu

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Protein binding
|
PDB id
|
|

|

|
4xuu
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Protein binding
|
 |
|
Title:
|
 |
The hsac2 domain from human phosphoinositide phosphatase sac2
|
|
Structure:
|
 |
Phosphatidylinositide phosphatase sac2. Chain: a, b, c, d. Fragment: unp residues 169-593. Synonym: inositol polyphosphate 5-phosphatase f,sac domain-containing inositol phosphatase 2,sac domain-containing phosphoinositide 5- phosphatase 2,hsac2. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: inpp5f, kiaa0966, sac2, mstp007, mstp047. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.62Å
|
R-factor:
|
0.250
|
R-free:
|
0.287
|
|
|
Authors:
|
 |
F.Hsu,Y.Mao
|
|
Key ref:
|
 |
F.Hsu
and
y.mao
Structure of the hsac2 domain at 2.62.
To be published,
.
|
 |
|
Date:
|
 |
|
26-Jan-15
|
Release date:
|
25-Feb-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9Y2H2
(SAC2_HUMAN) -
Phosphatidylinositide phosphatase SAC2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1132 a.a.
169 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.3.25
- inositol-phosphate phosphatase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
myo-Inositol Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a myo-inositol phosphate + H2O = myo-inositol + phosphate
|
 |
 |
 |
 |
 |
myo-inositol phosphate
|
+
|
H2O
|
=
|
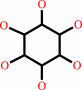 myo-inositol
myo-inositol
|
+
|
 phosphate
phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

