 |
PDBsum entry 4xom

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Unknown function
|
PDB id
|
|

|

|
4xom
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 206 a.a.
206 a.a.
|
 |

|

|

|

|

|

|

|
 193 a.a.
193 a.a.
|
 |

|

|

|

|

|

|

|
 178 a.a.
178 a.a.
|
 |

|

|
|
|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Unknown function
|
 |
|
Title:
|
 |
Coenzyme f420:l-glutamate ligase (fbib) from mycobacterium tuberculosis (c-terminal domain).
|
|
Structure:
|
 |
Coenzyme f420:l-glutamate ligase. Chain: a, b, c, d. Fragment: unp residues 245-448. Engineered: yes
|
|
Source:
|
 |
Mycobacterium tuberculosis (strain atcc 25618 / h37rv). Organism_taxid: 83332. Strain: h37rv. Cell_line: 25618. Gene: fbib, rv3262. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.233
|
R-free:
|
0.268
|
|
|
Authors:
|
 |
A.M.Rehan,G.Bashiri,H.M.Baker,E.N.Baker,C.J.Squire
|
|
Key ref:
|
 |
G.Bashiri
et al.
(2016).
Elongation of the Poly-γ-glutamate Tail of F420 Requires Both Domains of the F420:γ-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
J Biol Chem,
291,
6882-6894.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
16-Jan-15
|
Release date:
|
17-Feb-16
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P9WP79
(FBIB_MYCTU) -
Bifunctional F420 biosynthesis protein FbiB from Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
448 a.a.
206 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B, C, D:
E.C.1.3.8.17
- dehydro coenzyme F420 reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxidized coenzyme F420-0 + FMN + H+ = dehydro coenzyme F420-0 + FMNH2
|
 |
 |
 |
 |
 |
oxidized coenzyme F420-0
|
+
|
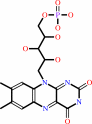 FMN
FMN
|
+
|
H(+)
|
=
|
dehydro coenzyme F420-0
|
+
|
 FMNH2
FMNH2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D:
E.C.6.3.2.31
- coenzyme F420-0:L-glutamate ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxidized coenzyme F420-0 + GTP + L-glutamate = oxidized coenzyme F420-1 + GDP + phosphate + H+
|
 |
 |
 |
 |
 |
oxidized coenzyme F420-0
|
+
|
 GTP
GTP
|
+
|
 L-glutamate
L-glutamate
|
=
|
oxidized coenzyme F420-1
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D:
E.C.6.3.2.34
- coenzyme F420-1:gamma-L-glutamate ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
oxidized coenzyme F420-1 + GTP + L-glutamate = oxidized coenzyme F420-2 + GDP + phosphate + H+
|
 |
 |
 |
 |
 |
oxidized coenzyme F420-1
|
+
|
 GTP
GTP
|
+
|
 L-glutamate
L-glutamate
|
=
|
oxidized coenzyme F420-2
|
+
|
 GDP
GDP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
291:6882-6894
(2016)
|
|
PubMed id:
|
|
|
|

|
| |
|
Elongation of the Poly-γ-glutamate Tail of F420 Requires Both Domains of the F420:γ-Glutamyl Ligase (FbiB) of Mycobacterium tuberculosis.
|
|
G.Bashiri,
A.M.Rehan,
S.Sreebhavan,
H.M.Baker,
E.N.Baker,
C.J.Squire.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cofactor F420is an electron carrier with a major role in the oxidoreductive
reactions ofMycobacterium tuberculosis, the causative agent of tuberculosis. A
γ-glutamyl ligase catalyzes the final steps of the F420biosynthesis pathway by
successive additions ofl-glutamate residues to F420-0, producing a
poly-γ-glutamate tail. The enzyme responsible for this reaction in archaea
(CofE) comprises a single domain and produces F420-2 as the major species. The
homologousM. tuberculosisenzyme, FbiB, is a two-domain protein and produces
F420with predominantly 5-7l-glutamate residues in the poly-γ-glutamate tail.
The N-terminal domain of FbiB is homologous to CofE with an annotated
γ-glutamyl ligase activity, whereas the C-terminal domain has sequence
similarity to an FMN-dependent family of nitroreductase enzymes. Here we
demonstrate that full-length FbiB adds multiplel-glutamate residues to F420-0in
vitroto produce F420-5 after 24 h; communication between the two domains is
critical for full γ-glutamyl ligase activity. We also present crystal
structures of the C-terminal domain of FbiB in apo-, F420-0-, and FMN-bound
states, displaying distinct sites for F420-0 and FMN ligands that partially
overlap. Finally, we discuss the features of a full-length structural model
produced by small angle x-ray scattering and its implications for the role of N-
and C-terminal domains in catalysis.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
| |

