 |
PDBsum entry 4x2q

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4x2q
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of human aldehyde dehydrogenase, aldh1a2
|
|
Structure:
|
 |
Retinal dehydrogenase 2. Chain: a, b, c, d. Synonym: raldh2,aldehyde dehydrogenase family 1 member a2, retinaldehyde-specific dehydrogenase type 2,raldh(ii). Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: aldh1a2, raldh2. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.94Å
|
R-factor:
|
0.268
|
R-free:
|
0.313
|
|
|
Authors:
|
 |
R.E.Stenkamp,I.Le Trong,J.K.Amory,J.Paik,A.S.Goldstein
|
|
Key ref:
|
 |
A.S.Goldstein
et al.
Synthesis and in vitro testing of bisdichloroacetyldi analogs for use as a reversible male contraceptive.
To be published,
.
|
 |
|
Date:
|
 |
|
26-Nov-14
|
Release date:
|
02-Dec-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O94788
(AL1A2_HUMAN) -
Retinal dehydrogenase 2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
518 a.a.
472 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.2.1.36
- retinal dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
retinal + NAD+ + H2O = retinoate + NADH + 2 H+
|
 |
 |
 |
 |
 |
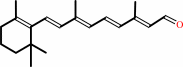 retinal
retinal
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 61.36% similarity
|
+
|
H2O
|
=
|
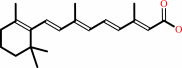 retinoate
retinoate
|
+
|
 NADH
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
 FAD
FAD
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

