 |
PDBsum entry 4wlm
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.62
- xylosyl alpha-1,3-xylosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-O-[alpha-D-xylosyl-(1->3)-beta-D-glucosyl]-L-seryl-[EGF-like domain protein] + UDP-alpha-D-xylose = 3-O-[alpha-D-xylosyl-(1->3)-alpha-D- xylosyl-(1->3)-beta-D-glucosyl]-L-seryl-[EGF-like domain protein] + UDP + H+
|
 |
 |
 |
 |
 |
3-O-[alpha-D-xylosyl-(1->3)-beta-D-glucosyl]-L-seryl-[EGF-like domain protein]
|
+
|
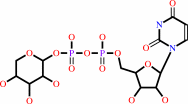 UDP-alpha-D-xylose
UDP-alpha-D-xylose
|
=
|
3-O-[alpha-D-xylosyl-(1->3)-alpha-D- xylosyl-(1->3)-beta-D-glucosyl]-L-seryl-[EGF-like domain protein]
|
+
|
 UDP
UDP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Chem Biol
11:847-854
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Notch-modifying xylosyltransferase structures support an SNi-like retaining mechanism.
|
|
H.Yu,
M.Takeuchi,
J.LeBarron,
J.Kantharia,
E.London,
H.Bakker,
R.S.Haltiwanger,
H.Li,
H.Takeuchi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
A major question remaining in glycobiology is how a glycosyltransferase (GT)
that retains the anomeric linkage of a sugar catalyzes the reaction. Xyloside
α-1,3-xylosyltransferase (XXYLT1) is a retaining GT that regulates Notch
receptor activation by adding xylose to the Notch extracellular domain. Here,
using natural acceptor and donor substrates and active Mus musculus XXYLT1, we
report a series of crystallographic snapshots along the reaction, including an
unprecedented natural and competent Michaelis reaction complex for retaining
enzymes. These structures strongly support the SNi-like reaction as the
retaining mechanism for XXYLT1. Unexpectedly, the epidermal growth factor-like
repeat acceptor substrate undergoes a large conformational change upon binding
to the active site, providing a structural basis for substrate specificity. Our
improved understanding of this retaining enzyme will accelerate the design of
retaining GT inhibitors that can modulate Notch activity in pathological
situations in which Notch dysregulation is known to cause cancer or
developmental disorders.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

