 |
PDBsum entry 4u72

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4u72
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase (a260g mutant)
|
|
Structure:
|
 |
Indoleamine 2,3-dioxygenase 1. Chain: a, b. Fragment: indoleamine 2,3-dioxygenase. Synonym: ido-1,indoleamine-pyrrole 2,3-dioxygenase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: ido1, ido, indo. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.188
|
R-free:
|
0.231
|
|
|
Authors:
|
 |
H.Sugimoto,M.Horitani,E.Kometani,Y.Shiro
|
|
Key ref:
|
 |
M.Horitani
et al.
Conformation and mobility of active site loop is crit for substrate binding and inhibition in human indolea 2,3-Dioxygenase.
To be published,
.
|
 |
|
Date:
|
 |
|
30-Jul-14
|
Release date:
|
02-Sep-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14902
(I23O1_HUMAN) -
Indoleamine 2,3-dioxygenase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
403 a.a.
375 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.52
- indoleamine 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
D-tryptophan + O2 = N-formyl-D-kynurenine
|
|
2.
|
L-tryptophan + O2 = N-formyl-L-kynurenine
|
|
 |
 |
 |
 |
 |
D-tryptophan
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
 O2
O2
|
=
|
 N-formyl-D-kynurenine
N-formyl-D-kynurenine
|
|
 |
 |
 |
 |
 |
L-tryptophan
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
 O2
O2
|
=
|
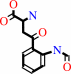 N-formyl-L-kynurenine
N-formyl-L-kynurenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

