 |
PDBsum entry 4ts3
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.1
- purine-nucleoside phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
a purine D-ribonucleoside + phosphate = a purine nucleobase + alpha- D-ribose 1-phosphate
|
|
2.
|
a purine 2'-deoxy-D-ribonucleoside + phosphate = a purine nucleobase + 2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine D-ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
purine nucleobase
|
+
|
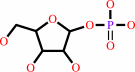 alpha- D-ribose 1-phosphate
alpha- D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
purine 2'-deoxy-D-ribonucleoside
|
+
|
phosphate
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
purine nucleobase
|
+
|
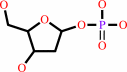 2-deoxy-alpha-D-ribose 1-phosphate
2-deoxy-alpha-D-ribose 1-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Sci Rep
8:15427
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallographic snapshots of ligand binding to hexameric purine nucleoside phosphorylase and kinetic studies give insight into the mechanism of catalysis.
|
|
Z.Štefanić,
M.Narczyk,
G.Mikleušević,
S.Kazazić,
A.Bzowska,
M.Luić.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Purine nucleoside phosphorylase (PNP) catalyses the cleavage of the glycosidic
bond of purine nucleosides using phosphate instead of water as a second
substrate. PNP from Escherichia coli is a homohexamer, build as a trimer of
dimers, and each subunit can be in two conformations, open or closed. This
conformational change is induced by the presence of phosphate substrate, and
very likely a required step for the catalysis. Closing one active site strongly
affects the others, by a yet unclear mechanism and order of events. Kinetic and
ligand binding studies show strong negative cooperativity between subunits.
Here, for the first time, we managed to monitor the sequence of nucleoside
binding to individual subunits in the crystal structures of the wild-type
enzyme, showing that first the closed sites, not the open ones, are occupied by
the nucleoside. However, two mutations within the active site,
Asp204Ala/Arg217Ala, are enough not only to significantly reduce the
effectiveness of the enzyme, but also reverse the sequence of the nucleoside
binding. In the mutant the open sites, neighbours in a dimer of those in the
closed conformation, are occupied as first. This demonstrates how important for
the effective catalysis of Escherichia coli PNP is proper subunit cooperation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

