 |
PDBsum entry 4s3d

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4s3d
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Ispg in complex with ppi
|
|
Structure:
|
 |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase. Chain: a. Synonym: 1-hydroxy-2-methyl-2-(e)-butenyl 4-diphosphate synthase. Engineered: yes
|
|
Source:
|
 |
Thermus thermophilus hb8. Organism_taxid: 300852. Strain: hb8/dsm 579. Atcc: 27634. Gene: ispg, ttha0305. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.144
|
R-free:
|
0.192
|
|
|
Authors:
|
 |
F.Quitterer,A.Frank,K.Wang,G.Rao,B.O'Dowd,J.Li,F.Guerra,S.Abdel- Azeim,A.Bacher,J.Eppinger,E.Oldfield,M.Groll
|
|
Key ref:
|
 |
F.Quitterer
et al.
(2015).
Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J Mol Biol,
427,
2220-2228.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
26-Jan-15
|
Release date:
|
29-Apr-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5SLI8
(ISPG_THET8) -
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (flavodoxin) from Thermus thermophilus (strain ATCC 27634 / DSM 579 / HB8)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
406 a.a.
402 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.17.7.3
- (E)-4-hydroxy-3-methylbut-2-enyl-diphosphate synthase (flavodoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2E)-4-hydroxy-3-methylbut-2-enyl diphosphate + oxidized [flavodoxin] + H2O + 2 H+ = 2-C-methyl-D-erythritol 2,4-cyclic diphosphate + reduced [flavodoxin]
|
 |
 |
 |
 |
 |
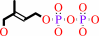 (E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate
(E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate
|
+
|
H(2)O
|
+
|
oxidized flavodoxin
|
=
|
 2-C-methyl-D-erythritol 2,4-cyclodiphosphate
2-C-methyl-D-erythritol 2,4-cyclodiphosphate
|
+
|
reduced flavodoxin
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
427:2220-2228
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
|
|
F.Quitterer,
A.Frank,
K.Wang,
G.Rao,
B.O'Dowd,
J.Li,
F.Guerra,
S.Abdel-Azeim,
A.Bacher,
J.Eppinger,
E.Oldfield,
M.Groll.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
IspG is the penultimate enzyme in non-mevalonate biosynthesis of the universal
terpene building blocks isopentenyl diphosphate and dimethylallyl diphosphate.
Its mechanism of action has been the subject of numerous studies but remained
unresolved due to difficulties in identifying distinct reaction intermediates.
Using a moderate reducing agent and an epoxide substrate analogue, we were now
able to trap and crystallographically characterize various stages in the
IspG-catalyzed conversion of 2-C-methyl-d-erythritol-2,4-cyclo-diphosphate into
(E)-1-hydroxy-2-methylbut-2-enyl-4-diphosphate. In addition, the enzyme's
structure was determined in complex with several inhibitors. These results,
combined with recent electron paramagnetic resonance data, allowed us to deduce
a detailed and complete IspG catalytic mechanism, which describes all stages
from initial ring opening to formation of
(E)-1-hydroxy-2-methylbut-2-enyl-4-diphosphate via discrete radical and
carbanion intermediates. The data presented in this article provide a guide for
the design of selective drugs against many prokaryotic and eukaryotic pathogens
to which the non-mevalonate pathway is essential for survival and virulence.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

