 |
PDBsum entry 4pxc
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
The crystal structure of atuah in complex with (s)-hydroxyglycine
|
|
Structure:
|
 |
Ureidoglycolate hydrolase. Chain: a, b. Fragment: unp residues 50-476. Synonym: atuah, allantoate amidohydrolase 2, atahh2, ureidoglycolate amidohydrolase. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Arabidopsis thaliana. Mouse-ear cress,thale-cress. Organism_taxid: 3702. Gene: uah, aah2, at5g43600, k9d7.10. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.89Å
|
R-factor:
|
0.169
|
R-free:
|
0.195
|
|
|
Authors:
|
 |
I.Shin,S.Rhee
|
|
Key ref:
|
 |
I.Shin
et al.
(2014).
Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
J Mol Biol,
426,
3028-3040.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
23-Mar-14
|
Release date:
|
23-Jul-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8VXY9
(UAH_ARATH) -
Ureidoglycolate hydrolase from Arabidopsis thaliana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
476 a.a.
423 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.1.116
- ureidoglycolate amidohydrolase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(S)-ureidoglycolate + H2O + 2 H+ = glyoxylate + 2 NH4+ + CO2
|
 |
 |
 |
 |
 |
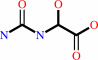 (S)-ureidoglycolate
(S)-ureidoglycolate
|
+
|
H2O
|
+
|
2
×
H(+)
|
=
|
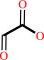 glyoxylate
glyoxylate
|
+
|
2
×
NH4(+)
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
426:3028-3040
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the substrate specificity of (s)-ureidoglycolate amidohydrolase and its comparison with allantoate amidohydrolase.
|
|
I.Shin,
K.Han,
S.Rhee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In plants, the ureide pathway is a metabolic route that converts the ring
nitrogen atoms of purine into ammonia via sequential enzymatic reactions,
playing an important role in nitrogen recovery. In the final step of the
pathway, (S)-ureidoglycolate amidohydrolase (UAH) catalyzes the conversion of
(S)-ureidoglycolate into glyoxylate and releases two molecules of ammonia as
by-products. UAH is homologous in structure and sequence with allantoate
amidohydrolase (AAH), an upstream enzyme in the pathway with a similar function
as that of an amidase but with a different substrate. Both enzymes exhibit
strict substrate specificity and catalyze reactions in a concerted manner,
resulting in purine degradation. Here, we report three crystal structures of
Arabidopsis thaliana UAH (bound with substrate, reaction intermediate, and
product) and a structure of Escherichia coli AAH complexed with allantoate.
Structural analyses of UAH revealed a distinct binding mode for each ligand in a
bimetal reaction center with the active site in a closed conformation. The
ligand directly participates in the coordination shell of two metal ions and is
stabilized by the surrounding residues. In contrast, AAH, which exhibits a
substrate-binding site similar to that of UAH, requires a larger active site due
to the additional ureido group in allantoate. Structural analyses and
mutagenesis revealed that both enzymes undergo an open-to-closed conformational
transition in response to ligand binding and that the active-site size and the
interaction environment in UAH and AAH are determinants of the substrate
specificities of these two structurally homologous enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

