 |
PDBsum entry 4onc
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.5.1.86
- trans,polycis-decaprenyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2Z,6E)-farnesyl diphosphate + 7 isopentenyl diphosphate = (2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E)-decaprenyl diphosphate + 7 diphosphate
|
 |
 |
 |
 |
 |
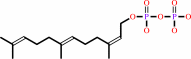 (2Z,6E)-farnesyl diphosphate
(2Z,6E)-farnesyl diphosphate
|
+
|
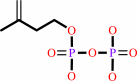 7
×
isopentenyl diphosphate
7
×
isopentenyl diphosphate
|
=
|
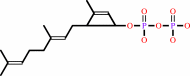 (2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E)-decaprenyl diphosphate
(2Z,6Z,10Z,14Z,18Z,22Z,26Z,30Z,34E)-decaprenyl diphosphate
|
+
|
 7
×
diphosphate
7
×
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.5.1.87
- ditrans,polycis-polyprenyl diphosphate synthase [(2E,6E)-farnesyl
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
n isopentenyl diphosphate + (2E,6E)-farnesyl diphosphate = a di-trans,poly-cis-polyprenyl diphosphate + n diphosphate
|
 |
 |
 |
 |
 |
 n
isopentenyl diphosphate
n
isopentenyl diphosphate
|
+
|
 7
×
(2E,6E)-farnesyl diphosphate
7
×
(2E,6E)-farnesyl diphosphate
|
=
|
di-trans,poly-cis-polyprenyl diphosphate
Bound ligand (Het Group name = )
matches with 44.19% similarity
|
+
|
 n
diphosphate
n
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Am Chem Soc
136:2892-2896
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
|
|
H.C.Chan,
X.Feng,
T.P.Ko,
C.H.Huang,
Y.Hu,
Y.Zheng,
S.Bogue,
C.Nakano,
T.Hoshino,
L.Zhang,
P.Lv,
W.Liu,
D.C.Crick,
P.H.Liang,
A.H.Wang,
E.Oldfield,
R.T.Guo.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We have obtained the structure of the bacterial diterpene synthase,
tuberculosinol/iso-tuberculosinol synthase (Rv3378c) from Mycobacterium
tuberculosis , a target for anti-infective therapies that block virulence factor
formation. This phosphatase adopts the same fold as found in the Z- or
cis-prenyltransferases. We also obtained structures containing the
tuberculosinyl diphosphate substrate together with one bisphosphonate
inhibitor-bound structure. These structures together with the results of
site-directed mutagenesis suggest an unusual mechanism of action involving two
Tyr residues. Given the similarity in local and global structure between Rv3378c
and the M. tuberculosis cis-decaprenyl diphosphate synthase (DPPS; Rv2361c), the
possibility exists for the development of inhibitors that target not only
virulence but also cell wall biosynthesis, based in part on the structures
reported here.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

