 |
PDBsum entry 4n2h
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Crystal structure of protein arginine deiminase 2 (d177a, 0 mm ca2+)
|
|
Structure:
|
 |
Protein-arginine deiminase type-2. Chain: a. Synonym: pad-h19, peptidylarginine deiminase ii, protein-arginine deiminase type ii. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: padi2, kiaa0994, pdi2. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.81Å
|
R-factor:
|
0.174
|
R-free:
|
0.211
|
|
|
Authors:
|
 |
D.J.Slade,X.Zhang,P.Fang,C.J.Dreyton,Y.Zhang,M.L.Gross,M.Guo, S.A.Coonrod,P.R.Thompson
|
|
Key ref:
|
 |
D.J.Slade
et al.
(2015).
Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem Biol,
10,
1043-1053.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
04-Oct-13
|
Release date:
|
04-Feb-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9Y2J8
(PADI2_HUMAN) -
Protein-arginine deiminase type-2 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
665 a.a.
639 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.3.15
- protein-arginine deiminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-arginyl-[protein] + H2O = L-citrullyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
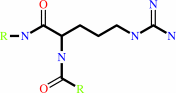 Protein L-arginine
Protein L-arginine
|
+
|
H(2)O
|
=
|
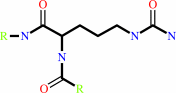 protein L-citrulline
protein L-citrulline
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acs Chem Biol
10:1043-1053
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
|
|
D.J.Slade,
P.Fang,
C.J.Dreyton,
Y.Zhang,
J.Fuhrmann,
D.Rempel,
B.D.Bax,
S.A.Coonrod,
H.D.Lewis,
M.Guo,
M.L.Gross,
P.R.Thompson.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Protein arginine deiminases (PADs) are calcium-dependent histone-modifying
enzymes whose activity is dysregulated in inflammatory diseases and cancer. PAD2
functions as an Estrogen Receptor (ER) coactivator in breast cancer cells via
the citrullination of histone tail arginine residues at ER binding sites.
Although an attractive therapeutic target, the mechanisms that regulate PAD2
activity are largely unknown, especially the detailed role of how calcium
facilitates enzyme activation. To gain insights into these regulatory processes,
we determined the first structures of PAD2 (27 in total), and through
calcium-titrations by X-ray crystallography, determined the order of binding and
affinity for the six calcium ions that bind and activate this enzyme. These
structures also identified several PAD2 regulatory elements, including a calcium
switch that controls proper positioning of the catalytic cysteine residue, and a
novel active site shielding mechanism. Additional biochemical and
mass-spectrometry-based hydrogen/deuterium exchange studies support these
structural findings. The identification of multiple intermediate calcium-bound
structures along the PAD2 activation pathway provides critical insights that
will aid the development of allosteric inhibitors targeting the PADs.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

