 |
PDBsum entry 4mw0

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Ligase/ligase inhibitor
|
PDB id
|
|

|

|
4mw0
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase/ligase inhibitor
|
 |
|
Title:
|
 |
Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-(2-hydroxyphenyl)urea (chem 1392)
|
|
Structure:
|
 |
Methionyl-tRNA synthetase. Chain: a, b. Fragment: unp residues 237-773. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Trypanosoma brucei. Organism_taxid: 5691. Gene: tb10.70.6470. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.185
|
R-free:
|
0.205
|
|
|
Authors:
|
 |
C.Y.Koh,J.E.Kim,A.B.Wetzel,W.J.De Van Der Schueren,S.Shibata,J.Liu, Z.Zhang,E.Fan,C.L.M.J.Verlinde,W.G.J.Hol
|
|
Key ref:
|
 |
C.Y.Koh
et al.
(2014).
Structures of Trypanosoma brucei methionyl-tRNA synthetase with urea-based inhibitors provide guidance for drug design against sleeping sickness.
Plos Negl Trop Dis,
8,
e2775.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
24-Sep-13
|
Release date:
|
30-Apr-14
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q38C91
(Q38C91_TRYB2) -
methionine--tRNA ligase from Trypanosoma brucei brucei (strain 927/4 GUTat10.1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
773 a.a.
531 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 7 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.10
- methionine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Met) + L-methionine + ATP = L-methionyl-tRNA(Met) + AMP + diphosphate
|
 |
 |
 |
 |
 |
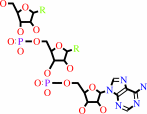 tRNA(Met)
tRNA(Met)
|
+
|
L-methionine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ATP
ATP
|
=
|
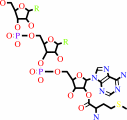 L-methionyl-tRNA(Met)
L-methionyl-tRNA(Met)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Plos Negl Trop Dis
8:e2775
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of Trypanosoma brucei methionyl-tRNA synthetase with urea-based inhibitors provide guidance for drug design against sleeping sickness.
|
|
C.Y.Koh,
J.E.Kim,
A.B.Wetzel,
W.J.de van der Schueren,
S.Shibata,
R.M.Ranade,
J.Liu,
Z.Zhang,
J.R.Gillespie,
F.S.Buckner,
C.L.Verlinde,
E.Fan,
W.G.Hol.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Methionyl-tRNA synthetase of Trypanosoma brucei (TbMetRS) is an important target
in the development of new antitrypanosomal drugs. The enzyme is essential,
highly flexible and displaying a large degree of changes in protein domains and
binding pockets in the presence of substrate, product and inhibitors. Targeting
this protein will benefit from a profound understanding of how its structure
adapts to ligand binding. A series of urea-based inhibitors (UBIs) has been
developed with IC50 values as low as 19 nM against the enzyme. The UBIs were
shown to be orally available and permeable through the blood-brain barrier, and
are therefore candidates for development of drugs for the treatment of late
stage human African trypanosomiasis. Here, we expand the structural diversity of
inhibitors from the previously reported collection and tested for their
inhibitory effect on TbMetRS and on the growth of T. brucei cells. The binding
modes and binding pockets of 14 UBIs are revealed by determination of their
crystal structures in complex with TbMetRS at resolutions between 2.2 Å to 2.9
Å. The structures show binding of the UBIs through conformational selection,
including occupancy of the enlarged methionine pocket and the auxiliary pocket.
General principles underlying the affinity of UBIs for TbMetRS are derived from
these structures, in particular the optimum way to fill the two binding pockets.
The conserved auxiliary pocket might play a role in binding tRNA. In addition, a
crystal structure of a ternary TbMetRS•inhibitor•AMPPCP complex indicates
that the UBIs are not competing with ATP for binding, instead are interacting
with ATP through hydrogen bond. This suggests a possibility that a general
'ATP-engaging' binding mode can be utilized for the design and development of
inhibitors targeting tRNA synthetases of other disease-causing pathogen.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

