 |
PDBsum entry 4mq4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase/transferase inhibitor
|
PDB id
|
|

|

|
4mq4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase/transferase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of hpnmt in complex with bisubstrate inhibitor n-(3- ((((2s,3s,4r,5r)-5-(6-amino-9h-purin-9-yl)-3,4- dihydroxytetrahydrofuran-2-yl)methyl)thio)propyl)-1,2,3,4- tetrahydroisoquinoline-3-carboxamide
|
|
Structure:
|
 |
Phenylethanolamine n-methyltransferase. Chain: a, b. Synonym: pnmtase, noradrenaline n-methyltransferase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: pnmt, pent. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.183
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
A.G.Bart,E.E.Scott
|
|
Key ref:
|
 |
A.G.Bart
and
e.e.scott
To be published.
To be published,
.
|
 |
|
Date:
|
 |
|
15-Sep-13
|
Release date:
|
15-Oct-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P11086
(PNMT_HUMAN) -
Phenylethanolamine N-methyltransferase from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
282 a.a.
264 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.28
- phenylethanolamine N-methyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Dopa Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
phenylethanolamine + S-adenosyl-L-methionine = N-methylphenylethanolamine + S-adenosyl-L-homocysteine + H+
|
 |
 |
 |
 |
 |
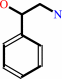 phenylethanolamine
phenylethanolamine
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
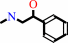 N-methylphenylethanolamine
N-methylphenylethanolamine
|
+
|
 S-adenosyl-L-homocysteine
S-adenosyl-L-homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 60.53% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

