 |
PDBsum entry 4mok

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4mok
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.10
- pyranose oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + O2 = 2-dehydro-D-glucose + H2O2
|
 |
 |
 |
 |
 |
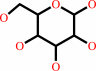 D-glucose
D-glucose
|
+
|
 O2
O2
|
=
|
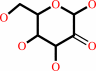 2-dehydro-D-glucose
2-dehydro-D-glucose
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Plos One
9:e86736
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for binding of fluorinated glucose and galactose to Trametes multicolor pyranose 2-oxidase variants with improved galactose conversion.
|
|
T.C.Tan,
O.Spadiut,
R.Gandini,
D.Haltrich,
C.Divne.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Each year, about six million tons of lactose are generated from liquid whey as
industrial byproduct, and optimally this large carbohydrate waste should be used
for the production of value-added products. Trametes multicolor pyranose
2-oxidase (TmP2O) catalyzes the oxidation of various monosaccharides to the
corresponding 2-keto sugars. Thus, a potential use of TmP2O is to convert the
products from lactose hydrolysis, D-glucose and D-galactose, to more valuable
products such as tagatose. Oxidation of glucose is however strongly favored over
galactose, and oxidation of both substrates at more equal rates is desirable.
Characterization of TmP2O variants (H450G, V546C, H450G/V546C) with improved
D-galactose conversion has been given earlier, of which H450G displayed the best
relative conversion between the substrates. To rationalize the changes in
conversion rates, we have analyzed high-resolution crystal structures of the
aforementioned mutants with bound 2- and 3-fluorinated glucose and galactose.
Binding of glucose and galactose in the productive 2-oxidation binding mode is
nearly identical in all mutants, suggesting that this binding mode is
essentially unaffected by the mutations. For the competing glucose binding mode,
enzyme variants carrying the H450G replacement stabilize glucose as the
α-anomer in position for 3-oxidation. The backbone relaxation at position 450
allows the substrate-binding loop to fold tightly around the ligand. V546C
however stabilize glucose as the β-anomer using an open loop conformation.
Improved binding of galactose is enabled by subtle relaxation effects at key
active-site backbone positions. The competing binding mode for galactose
2-oxidation by V546C stabilizes the β-anomer for oxidation at C1, whereas H450G
variants stabilize the 3-oxidation binding mode of the galactose α-anomer. The
present study provides a detailed description of binding modes that rationalize
changes in the relative conversion rates of D-glucose and D-galactose and can be
used to refine future enzyme designs for more efficient use of
lactose-hydrolysis byproducts.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

