 |
PDBsum entry 4mhq
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Replication
|
 |
|
Title:
|
 |
Crystal structure of human primase catalytic subunit
|
|
Structure:
|
 |
DNA primase small subunit. Chain: a. Fragment: full length. Synonym: DNA primase 49 kda subunit, p49. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: prim1. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.179
|
R-free:
|
0.233
|
|
|
Authors:
|
 |
K.R.Park,J.Y.An,Y.Lee,H.S.Youn,J.G.Lee,J.Y.Kang,T.G.Kim,J.J.Lim, S.H.Eom,J.Wang
|
|
Key ref:
|
 |
K.R.Park
et al.
Crystal structure of human primase catalytic subunit.
To be published,
.
|
 |
|
Date:
|
 |
|
30-Aug-13
|
Release date:
|
03-Sep-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P49642
(PRI1_HUMAN) -
DNA primase small subunit from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
420 a.a.
400 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.7.102
- Dna primase aep.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
ssDNA + n NTP = ssDNA/pppN(pN)n-1 hybrid + (n-1) diphosphate
|
|
2.
|
ssDNA + n dNTP = ssDNA/pppdN(pdN)n-1 hybrid + (n-1) diphosphate
|
|
 |
 |
 |
 |
 |
ssDNA
|
+
|
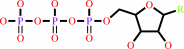 n
NTP
n
NTP
|
=
|
ssDNA/pppN(pN)n-1 hybrid
|
+
|
(n-1) diphosphate
|
|
 |
 |
 |
 |
 |
ssDNA
|
+
|
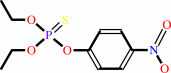 n
dNTP
n
dNTP
|
=
|
ssDNA/pppdN(pdN)n-1 hybrid
|
+
|
(n-1) diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

