 |
PDBsum entry 4m0c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4m0c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
The crystal structure of a fmn-dependent nadh-azoreductase from bacillus anthracis str. Ames ancestor in complex with fmn.
|
|
Structure:
|
 |
Fmn-dependent nadh-azoreductase 1. Chain: a, b. Synonym: azo-dye reductase 1, fmn-dependent nadh-azo compound oxidoreductase 1. Engineered: yes
|
|
Source:
|
 |
Bacillus anthracis. Anthrax,anthrax bacterium. Organism_taxid: 261594. Strain: ames ancestor. Gene: azor1,bas0908, ba_0966, gbaa_0966. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
2.07Å
|
R-factor:
|
0.178
|
R-free:
|
0.224
|
|
|
Authors:
|
 |
K.Tan,M.Gu,K.Kwon,W.F.Anderson,A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
K.Tan
et al.
The crystal structure of a fmn-Dependent nadh-Azoredu from bacillus anthracis str. Ames ancestor in complex fmn..
To be published,
.
|
 |
|
Date:
|
 |
|
01-Aug-13
|
Release date:
|
14-Aug-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q81UB2
(AZOR1_BACAN) -
FMN-dependent NADH:quinone oxidoreductase 1 from Bacillus anthracis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
220 a.a.
212 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.6.5.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.1.7.1.17
- FMN-dependent NADH-azoreductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N,N-dimethyl-1,4-phenylenediamine + anthranilate + 2 NAD+ = 2-(4-dimethylaminophenyl)diazenylbenzoate + 2 NADH + 2 H+
|
 |
 |
 |
 |
 |
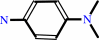 N,N-dimethyl-1,4-phenylenediamine
N,N-dimethyl-1,4-phenylenediamine
|
+
|
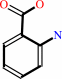 anthranilate
anthranilate
|
+
|
 2
×
NAD(+)
2
×
NAD(+)
|
=
|
2-(4-dimethylaminophenyl)diazenylbenzoate
|
+
|
 2
×
NADH
2
×
NADH
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FMN
|
 |
 |
 |
 |
 |
FMN
Bound ligand (Het Group name =
FMN)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

