 |
PDBsum entry 4lza
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of adenine phosphoribosyltransferase from thermoanaerobacter pseudethanolicus atcc 33223, nysgrc target 029700.
|
|
Structure:
|
 |
Adenine phosphoribosyltransferase. Chain: a, b. Synonym: aprt. Engineered: yes
|
|
Source:
|
 |
Thermoanaerobacter pseudethanolicus. Organism_taxid: 340099. Strain: atcc 33223. Gene: 166856274, apt, teth39_1027. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.84Å
|
R-factor:
|
0.205
|
R-free:
|
0.228
|
|
|
Authors:
|
 |
V.N.Malashkevich,R.Bhosle,R.Toro,B.Hillerich,A.Gizzi,S.Garforth, A.Kar,M.K.Chan,J.Lafluer,H.Patel,B.Matikainen,S.Chamala,S.Lim, A.Celikgil,G.Villegas,B.Evans,J.Love,A.Fiser,K.Khafizov,R.Seidel, J.B.Bonanno,S.C.Almo,New York Structural Genomics Research Consortium (Nysgrc)
|
|
Key ref:
|
 |
V.N.Malashkevich
et al.
Crystal structure of adenine phosphoribosyltransferas thermoanaerobacter pseudethanolicus atcc 33223, Nysgr target 029700..
To be published,
.
|
 |
|
Date:
|
 |
|
31-Jul-13
|
Release date:
|
14-Aug-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
B0K969
(APT_THEP3) -
Adenine phosphoribosyltransferase from Thermoanaerobacter pseudethanolicus (strain ATCC 33223 / 39E)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
173 a.a.
168 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.7
- adenine phosphoribosyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Ribose activation
|
 |
 |
 |
 |
 |
Reaction:
|
 |
AMP + diphosphate = 5-phospho-alpha-D-ribose 1-diphosphate + adenine
|
 |
 |
 |
 |
 |
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
=
|
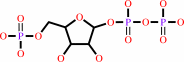 5-phospho-alpha-D-ribose 1-diphosphate
5-phospho-alpha-D-ribose 1-diphosphate
|
+
|
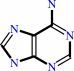 adenine
adenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

