 |
PDBsum entry 4l6h
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.14
- 5-methyltetrahydropteroyltriglutamate--homocysteine S-methyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5-methyltetrahydropteroyltri-L-glutamate + L-homocysteine = tetrahydropteroyltri-L-glutamate + L-methionine
|
 |
 |
 |
 |
 |
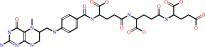 5-methyltetrahydropteroyltri-L-glutamate
5-methyltetrahydropteroyltri-L-glutamate
|
+
|
L-homocysteine
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
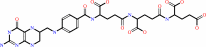 tetrahydropteroyltri-L-glutamate
tetrahydropteroyltri-L-glutamate
|
+
|
L-methionine
Bound ligand (Het Group name = )
matches with 56.60% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Zn(2+)
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
426:1839-1847
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural analysis of a fungal methionine synthase with substrates and inhibitors.
|
|
D.Ubhi,
G.Kago,
A.F.Monzingo,
J.D.Robertus.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The cobalamin-independent methionine synthase from Candida albicans, known as
Met6p, is a 90-kDa enzyme that consists of two (βα)8 barrels. The active site
is located between the two domains and has binding sites for a zinc ion and
substrates l-homocysteine and 5-methyl-tetrahydrofolate-glutamate3. Met6p
catalyzes transfer of the methyl group of 5-methyl-tetrahydrofolate-glutamate3
to the l-homocysteine thiolate to generate methionine. Met6p is essential for
fungal growth, and we currently pursue it as an antifungal drug design target.
Here we report the binding of l-homocysteine, methionine, and several folate
analogs. We show that binding of l-homocysteine or methionine results in
conformational rearrangements at the amino acid binding pocket, moving the
catalytic zinc into position to activate the thiol group. We also map the folate
binding pocket and identify specific binding residues, like Asn126, whose
mutation eliminates catalytic activity. We also report the development of a
robust fluorescence-based activity assay suitable for high-throughput screening.
We use this assay and an X-ray structure to characterize methotrexate as a weak
inhibitor of fungal Met6p.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

