 |
PDBsum entry 4l41

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Calcium binding protein/transferase
|
PDB id
|
|

|

|
4l41
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Calcium binding protein/transferase
|
 |
|
Title:
|
 |
Human lactose synthase: a 2:1 complex between human alpha-lactalbumin and human beta1,4-galactosyltransferase
|
|
Structure:
|
 |
Alpha-lactalbumin. Chain: a, b. Fragment: unp residues 19-142. Synonym: lactose synthase b protein, lysozyme-like protein 7. Engineered: yes. Beta-1,4-galactosyltransferase 1. Chain: c. Fragment: unp residues 126-398. Synonym: beta-1,4-galtase 1, beta4gal-t1, b4gal-t1.
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: 4-galactosyltransferase, beta1, lalba, lyzl7. Expressed in: escherichia coli. Expression_system_taxid: 511693. Gene: alpha-lactalbumin, b4galt1, ggtb2.
|
|
Resolution:
|
 |
|
2.70Å
|
R-factor:
|
0.190
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
B.Ramakrishnan,P.K.Qasba
|
|
Key ref:
|
 |
B.Ramakrishnan
and
p.k.qasba
Crystal structure of human lactose synthase forms a n 1:2 complex between beta1,4galactosyltransferase 1 an alpha-Lactalbumin proteins.
To be published,
.
|
 |
|
Date:
|
 |
|
07-Jun-13
|
Release date:
|
02-Oct-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chain C:
E.C.2.4.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chain C:
E.C.2.4.1.22
- lactose synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + UDP-alpha-D-galactose = lactose + UDP + H+
|
 |
 |
 |
 |
 |
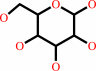 D-glucose
D-glucose
|
+
|
UDP-alpha-D-galactose
|
=
|
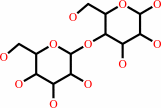 lactose
lactose
|
+
|
 UDP
UDP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chain C:
E.C.2.4.1.275
- neolactotriaosylceramide beta-1,4-galactosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a beta-D-GlcNAc-(1->3)-beta-D-Gal-(1->4)-beta-D-Glc-(1<->1)- Cer(d18:1(4E)) + UDP-alpha-D-galactose = a neolactoside nLc4Cer(d18:1(4E)) + UDP + H+
|
 |
 |
 |
 |
 |
beta-D-GlcNAc-(1->3)-beta-D-Gal-(1->4)-beta-D-Glc-(1<->1)- Cer(d18:1(4E))
|
+
|
UDP-alpha-D-galactose
|
=
|
neolactoside nLc4Cer(d18:1(4E))
|
+
|
 UDP
UDP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chain C:
E.C.2.4.1.38
- beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an N-acetyl-beta-D-glucosaminyl derivative + UDP-alpha-D-galactose = a beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl derivative + UDP + H+
|
 |
 |
 |
 |
 |
N-acetyl-beta-D-glucosaminyl derivative
|
+
|
UDP-alpha-D-galactose
|
=
|
beta-D-galactosyl-(1->4)-N-acetyl-beta-D-glucosaminyl derivative
|
+
|
 UDP
UDP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chain C:
E.C.2.4.1.90
- N-acetyllactosamine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N-acetyl-D-glucosamine + UDP-alpha-D-galactose = beta-D-galactosyl- (1->4)-N-acetyl-D-glucosamine + UDP + H+
|
 |
 |
 |
 |
 |
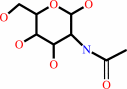 N-acetyl-D-glucosamine
N-acetyl-D-glucosamine
|
+
|
UDP-alpha-D-galactose
|
=
|
beta-D-galactosyl- (1->4)-N-acetyl-D-glucosamine
|
+
|
 UDP
UDP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

