 |
PDBsum entry 4jii

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/oxidoreductase inhibitor
|
PDB id
|
|

|

|
4jii
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/oxidoreductase inhibitor
|
 |
|
Title:
|
 |
Crystal structure of akr1b10 complexed with NADP+ and zopolrestat
|
|
Structure:
|
 |
Aldo-keto reductase family 1 member b10. Chain: x. Synonym: arl-1, aldose reductase-like, aldose reductase-related protein, arp, harp, small intestine reductase, si reductase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: akr1b10, akr1b11. Expressed in: escherichia coli. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
2.20Å
|
R-factor:
|
0.172
|
R-free:
|
0.218
|
|
|
Authors:
|
 |
L.Zhang,X.Zheng,H.Zhang,Y.Zhao,K.Chen,J.Zhai,X.Hu,Structural Genomics Consortium (Sgc)
|
|
Key ref:
|
 |
L.Zhang
et al.
(2013).
Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: role of Trp112 (Trp111).
Febs Lett,
587,
3681-3686.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
06-Mar-13
|
Release date:
|
23-Oct-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
O60218
(AK1BA_HUMAN) -
Aldo-keto reductase family 1 member B10 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
316 a.a.
318 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.300
- NADP-retinol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
all-trans-retinol + NADP+ = all-trans-retinal + NADPH + H+
|
 |
 |
 |
 |
 |
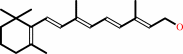 all-trans-retinol
all-trans-retinol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
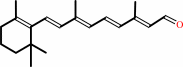 all-trans-retinal
all-trans-retinal
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.54
- allyl-alcohol dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
allyl alcohol + NADP+ = acrolein + NADPH + H+
|
 |
 |
 |
 |
 |
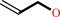 allyl alcohol
allyl alcohol
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
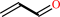 acrolein
acrolein
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Febs Lett
587:3681-3686
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: role of Trp112 (Trp111).
|
|
L.Zhang,
H.Zhang,
Y.Zhao,
Z.Li,
S.Chen,
J.Zhai,
Y.Chen,
W.Xie,
Z.Wang,
Q.Li,
X.Zheng,
X.Hu.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The antineoplastic target aldo-keto reductase family member 1B10 (AKR1B10) and
the critical polyol pathway enzyme aldose reductase (AKR1B1) share high
structural similarity. Crystal structures reported here reveal a surprising
Trp112 native conformation stabilized by a specific Gln114-centered hydrogen
bond network in the AKR1B10 holoenzyme, and suggest that AKR1B1 inhibitors could
retain their binding affinities toward AKR1B10 by inducing Trp112 flip to result
in an "AKR1B1-like" active site in AKR1B10, while selective AKR1B10
inhibitors can take advantage of the broader active site of AKR1B10 provided by
the native Trp112 side-chain orientation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

