 |
PDBsum entry 4jek

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4jek
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Structure of dibenzothiophene monooxygenase (dszc) from rhodococcus erythropolis
|
|
Structure:
|
 |
Dibenzothiophene desulfurization enzymE C. Chain: a, b, c, d, e, f, g, h. Synonym: dbt sulfur dioxygenase. Engineered: yes
|
|
Source:
|
 |
Rhodococcus sp.. Organism_taxid: 54064. Strain: igts8. Gene: soxc, dszc. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.209
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
L.Zhang,X.L.Duan,D.M.Zhou,X.Li
|
|
Key ref:
|
 |
X.Duan
et al.
(2013).
Crystallization and preliminary structural analysis of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis.
Acta Crystallogr Sect F Struct Biol Cryst Commun,
69,
597-601.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
27-Feb-13
|
Release date:
|
18-Sep-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q0ZIH5
(DSZC3_RHOER) -
Dibenzothiophene monooxygenase from Rhodococcus erythropolis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
417 a.a.
399 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 7 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.14.21
- dibenzothiophene monooxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dibenzothiophene + 2 FMNH2 + 2 O2 = dibenzothiophene 5,5-dioxide + 2 FMN + 2 H2O + 2 H+
|
 |
 |
 |
 |
 |
dibenzothiophene
|
+
|
 2
×
FMNH2
2
×
FMNH2
|
+
|
 2
×
O2
2
×
O2
|
=
|
dibenzothiophene 5,5-dioxide
|
+
|
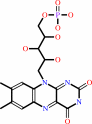 2
×
FMN
2
×
FMN
|
+
|
2
×
H2O
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
69:597-601
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystallization and preliminary structural analysis of dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis.
|
|
X.Duan,
L.Zhang,
D.Zhou,
K.Ji,
T.Ma,
W.Shui,
G.Li,
X.Li.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Dibenzothiophene (DBT) and its derivatives are typical sulfur compounds found in
fossil fuels. These compounds show resistance to the hydrodesulfurization
treatment that is commonly used in industry. Dibenzothiophene monooxygenase
(DszC) is responsible for the oxidation of DBT, which is the first and the
rate-limiting step in the DBT enzymatic desulfurization 4S pathway. In this
study, the crystal structure of DszC from Rhodococcus erythropolis DS-3 is
reported. The crystal of native DszC belonged to space group P1, with unit-cell
parameters a = 96.16, b = 96.27, c = 98.56 Å, α = 81.03, β = 67.57, γ =
85.84°. To determine the phase, SAD X-ray diffraction data were collected from
a SeMet-derivative DszC crystal, which also belonged to space group P1, with
unit-cell parameters a = 95.379, b = 95.167, c = 94.891 Å, α = 87.046, β =
70.536, γ = 79.738°. Further structural analysis of DszC is in progress.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

